17 December 2020: Review Articles
Current Status of Diagnostic Testing for SARS-CoV-2 Infection and Future Developments: A Review
Jing Gao 12ABCEF , Lei Quan 1AEF*DOI: 10.12659/MSM.928552
Med Sci Monit 2020; 26:e928552
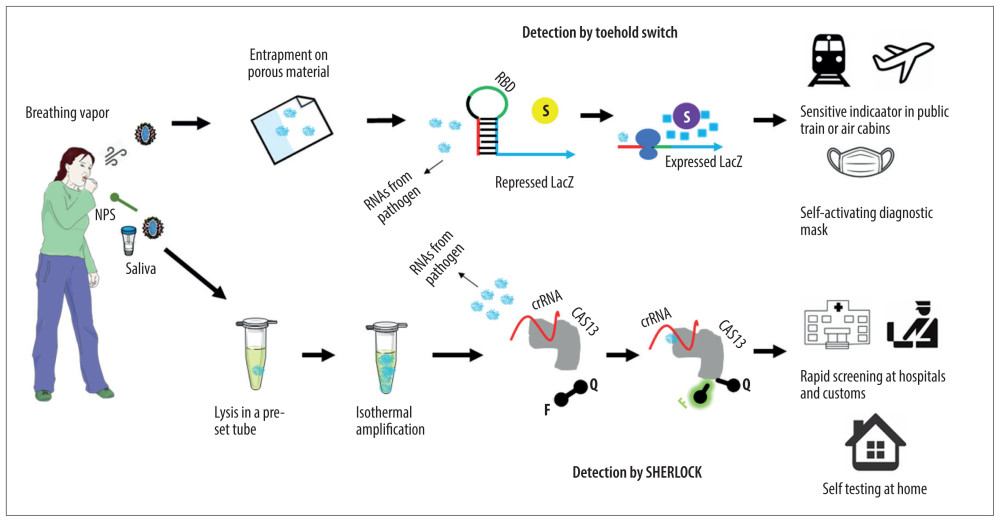
Figure 1 Novel RNA sensing techniques and their potential applications for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-COV-2) infection. On-site and rapid detection of emerging pathogens is feasible with the recent advances in molecular diagnostics. In a Toehold Switch system, virus in the breathing vapor of patients is attached by porous materials embedded with cell-free gene expression systems. The secondary RNA structure repressing the expression of LacZ consists of a sensing sequence (red), a linker sequence (black), a ribosome-binding domain (RBD, green), and an antisense sensoring sequence (blue). Sequence-specific binding of viral RNA to the sensoring sequence releases the secondary structure and lead to expression of LacZ (blue squares), which cleaves the substrate chlorophenol red-b-D-glucopyranoside (S, yellow) to produce a substrate of a visible purple color (S, purple). On a specific high-sensitivity enzymatic reporter unlocking (SHERLOCK) platform, clinical samples are collected by trained professionals or patients themselves following simple instruction. Collected samples are lysed (and therefore sterilized) and stabilized, amplified through an optional isothermal amplification process, and tested by a recently developed in vitro assay. Viral RNA can pair with the CRISPR RNA (crRNA) in a sequence-specific manner to activate robust and nonspecific single-stranded DNA trans-cleavage, which releases fluorescent signals (F) from their quenchers (Q) linked by single-stranded DNA. The fluorescent signal can be also replaced with a colorimetric visual readout on a lateral flow strip. Potential applications of these novel techniques for the on-site and rapid detection of emerging novel pathogens such as SARS-COV-2 are also depicted.


