08 October 2020: Clinical Research
Genome-Scale Expression Pattern of Long Non-Coding RNAs in Chinese Uyghur Patients with Parkinson’s Disease
Dan Wang1ABE, Hua Gao2BDF, Yanxia Li3CDF, Sen Jiang1CDG, Yuxuan Yong1DEF, Xinling Yang1AFG*DOI: 10.12659/MSM.925888
Med Sci Monit 2020; 26:e925888
Abstract
BACKGROUND: Long non-coding RNAs (lncRNAs) are transcripts thought to regulate gene expression at the post-transcriptional level. Some lncRNAs are associated with Parkinson’s disease (PD) and participate in pathological processes of PD. The incidence of PD is relatively high in members of the Uyghur minority living in Xingjiang province of China. This study measured the expression of lncRNAs in the peripheral blood cells of Chinese Uyghur individuals with and without PD and analyzed the possible function of these lncRNAs in the development of PD.
MATERIAL AND METHODS: Peripheral blood samples were collected from 55 Uyghur patients with PD and 55 healthy volunteers. Total RNA was extracted, and the levels of expression of whole-genome lncRNAs and mRNAs in 10 samples (5 PD and 5 controls) were determined by microarray method. The expression levels of lncRNAs in all 100 subjects were determined by qRT-PCR. The lncRNA expression profiles of PD patients were determined based on lncRNA microarray chip analysis, and differentially expressed lncRNAs were identified. The results of chip analysis were confirmed in a large clinical cohort.
RESULTS: Comparison of subjects with and without PD identified 32 significantly up-regulated and 18 significantly down-regulated lncRNAs in the PD group. GO analysis showed that mRNAs encoding proteins involved in the regulation of biological processes were differentially expressed, with the inflammatory immune response being the most significantly related pathway.
CONCLUSIONS: The expression of lncRNAs in peripheral blood differed significantly in PD patients and controls. These differentially expressed lncRNAs may play a role in the development of PD.
Keywords: Microarray Analysis, Asians, Gene Expression Regulation, Genome-Wide Association Study
Background
Parkinson’s disease (PD) is a common neurodegenerative disease, characterized by resting tremors, stiffness, bradykinesia and postural instability, as well as non-motor symptoms such as psychosis, sensory symptoms, autonomic dysfunction, and sleep disturbances. The main pathological changes of PD are the degeneration of dopamine neurons in the dense substantia nigra of the midbrain and the subsequent depletion of striatal dopamine [1,2]. Various epidemiological and experimental investigations have shown that aging, genetic factors and environmental toxins synergistically participate in degenerative damage to dopaminergic neurons [3–5]. The slow progression of PD and the continuous deterioration of clinical characteristics lead to disability and related complications. Identifying new biological information or biomarkers can better explore the pathogenesis of PD, improve its early diagnosis and identify potential therapeutic targets.
Long non-coding RNAs (lncRNAs) are a recently discovered group of RNA molecules, ranging in length from 200 bp to 10 kbp, which are thought to regulate gene expression at the post-transcriptional level, including in PD [6,7]. Although lncRNAs lack the ability to encode any proteins, they have crucial regulatory potential in processing proteins during many biological processes [8–10]. LncRNAs may play important roles in the pathological changes of PD, including in gene transcription, DNA methylation, post-transcriptional processes, epigenetic modification, direct protein binding and regulation of protein functions [11–13].
The Uyghur constitute an ethnic minority within China. Most Ughyur people live in Xinjiang Province, on the northwestern border of China. These people have a lifestyle and dietary habits different from those of Han Chinese, as well as a relatively high incidence of PD [14,15]. Few epidemiological studies have assessed PD in the Uyghur population. The present study assessed the expression of lncRNAs in Uyghur individuals with and without PD and analyzed the possible biological functions of these lncRNAs in the development of PD.
Material and Methods
RNA EXTRACTION:
Total RNA was extracted from peripheral blood cells using mirVana extraction kits (Ambion, Austin, TX, USA), and purified by QIAGEN RNeasy® kits [17–19]. RNA 6000 Nanochip Lab-on-a-Chip kits and the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA) were used to detect RNA integrity by capillary electrophoresis. Only RNA samples with RNA integrity values >6 were further analyzed. For chip preparation, total RNA samples were amplified in vitro and fluorescently labeled, with the labeling, hybridization, and scanning of the chip completed by Beijing Boao Biological Co. Ltd.
LNCRNA AND MRNA ANALYSIS:
Human LncRNA Array V4, 4×180k Agilent lncRNA was used for analysis, based on the latest information on lncRNAs in the GENCODE/ENSEMBL database, LNCipedia, the human LincRNA Catalog [20], the ncRNA expression database (NRED), and the RefSeq and USCS databases. This array allowed lncRNAs and mRNAs to be analyzed simultaneously, and their correlations determined. LncRNA expression profiling analyses were completed by Beijing Boao Biotechnology Co., Ltd.
GeneSpring software V13.0 (Agilent) was used for data aggregation, standardization and quality control analysis of lncRNA and mRNA array data.
LNCRNA RELATED FUNCTIONAL ANALYSES:
The potential functions of lncRNAs were predicted based on related cis- and trans-mRNAs. The regulated target gene was selected and sequences located 10 kb upstream and downstream of the coding gene position were subjected to Gene Ontology (GO) analysis to predict the biological significance of the target genes. Target genes were mapped to the Kyoto Encyclopedia of Genes and Genomes (KEGG) advanced database resource, and the main pathways of differentially expressed genes were identified based on KEGG results [21]. The lncRNA/mRNA co-expression networks were constructed based on Pearson correlation coefficients not less than 0.99 [22].
QRT-PCR:
Total RNA was extracted and reverse transcribed to cDNA using RNeasy Mini kits (Fermentas, K1622). The cDNA was analyzed by quantitative real-time polymerase chain reaction (qRT-PCR) using Power SYBR Green PCR Master (Applied Biosystems, Foster City, CA USA) and the primer sequences shown in Table 1. The expression of each was normalized to that of GAPDH mRNA in the same sample, and fold change (FC) of target gene expression in the experimental group relative to the control group calculated using the 2−ΔΔCT method.
STATISTICAL ANALYSIS:
All statistical data were analyzed with SPSS Statistics 18.0 software. Qualitative variables were compared by Pearson’s Chi-squared test. The normality of quantitative variables was determined. Normally distributed variables were expressed as mean±standard deviations (SD) and compared by Student’s t-tests, whereas non-normally distributed variables were reported as median (interquartile range [IQR]) and compared by Mann-Whitney U tests.
Results
CLINICAL CHARACTERISTICS:
This study included a total of 55 patients with PD and 55 age-matched healthy controls. Assessments of their clinical characteristics showed no significant differences (Table 2).
DIFFERENTIALLY EXPRESSED LNCRNAS IN PD PATIENTS AND HEALTHY CONTROLS:
Analysis of lncRNAs in samples from 5 Uyghur PD patients and 5 age-matched Uyghur healthy controls identified 50 differentially expressed lncRNAs with FC ≥2.0 and P <0.05. Of these 50 lncRNAs, 32 were up-regulated and 18 were down-regulated in PD patients relative to healthy controls (Table 3).
The relationships of lncRNAs between Uyghur PD and healthy controls were analyzed by hierarchical clustering [23,24]. The 2 groups of hierarchical clusters showed different expression profiles of lncRNAs (Figure 1A). The clustering of the same group of samples indicated consistent gene expression trends. The scatter plots and volcano plots provide a visual representation of the differences in lncRNA expression between the 2 groups (Figure 1B–1C).
FUNCTIONAL ANNOTATION OF LNCRNAS IN PD:
The potential function of lncRNAs was evaluated by annotation of co-expressed mRNAs. Ninety-seven differentially expressed mRNAs, with FC ≥2.0 and P<0.05 were identified, with 65 up-regulated and 32 down-regulated in PD patients relative to healthy controls (Figure 2A). The top 25 differentially expressed mRNAs are listed in Table 4. The scatter plots and volcano plots showed clear differences in expression of mRNAs between the 2 groups (Figure 2B–2C).
GO annotation showed that the top 5 terms related to biological processes in PD patients included: (1) cellular processes, (2) single-organism processes, (3) biological regulation, (4) regulation of biological processes, and (5) responses to stimuli. The GO terms most significantly associated with cellular components in PD patients included: (1) cells, (2) parts of cells, (3) organelles, (4) membranes, and (5) parts of membranes.
The GO terms most significantly associated with molecular function in PD patients included: (1) binding, (2) catalytic activity, (3) molecular transduction, (4) regulation of molecular function, and (5) nucleic acid binding transcription factor (Figure 3).
KEGG pathway analysis showed that the most enriched pathways corresponding to PD-related lncRNA disorders included: Cytokine-cytokine receptor interactions, chemokine receptors binding to chemokines, natural killer cell-mediated cytotoxicity, immunoregulatory interactions between lymphoid and non-lymphoid cells, and the NF-kappa B signaling pathway (Figure 4).
The most significantly enriched disease terms included: (1) immune system diseases, (2) allergies and autoimmune diseases, (3) gastric cancer, somatic, (4) common variable immunodeficiency, and (5) primary immunodeficiency (Figure 5).
LNCRNA-MRNA NETWORK ANALYSIS:
Based on Pearson correlation coefficients not less than 0.99, a co-expression network of differentially expressed lncRNAs and mRNAs was constructed (Figure 6).
VALIDATION OF LNCRNA BY QRT-PCR:
To verify the results of microarray analysis of lncRNAs expression, the levels of expression of 3 randomly selected lncRNAs of 50 Uyghur PD patients and 50 healthy controls were evaluated by qRT-PCR (Table 5). These results were consistent with those from microarray analysis.
Discussion
PD is a typical progressive neurodegenerative disease with a high prevalence worldwide. Although the pathogenesis of PD remains unclear, genetic factors are involved [25]. LncRNAs were shown to be involved in various neurodegenerative diseases, such as PD, Huntington’s disease, Alzheimer’s disease (AD), and spinocerebellar ataxia [26–28].
LncRNAs have been linked to the occurrence and development of PD. An analysis of the levels of expression of lncRNAs in 30 brain specimens from 20 PD patients and 10 controls found that 5 lncRNAs were significantly differentially expressed in these samples. Interestingly, analysis of the levels of expression of lncRNAs and disease stages showed that changes in lncRNA expression can be detected in patients with early PD, suggesting that lncRNA dysregulation may have occurred before PD [29]. Analysis of brain nigra tissue samples from 11 PD patients and 14 normal controls showed obvious changes in 87 lncRNAs. Among them, lncRNA AL049437 may trigger the development of PD, whereas lncRNA AK021630 may inhibit its occurrence [30]. Whole-transcriptome RNA sequencing technology has been used to determine all transcripts encoding proteins and lncRNAs in peripheral blood leukocytes of PD patients before and after deep brain stimulation (DBS). A comparison with healthy controls identified associations between expression of lncRNAs and selective PD-induced changes. Of the more than 6000 lncRNAs detected, 13 showed PD-induced changes, with 4 experiencing reverse changes after DBS [12]. Previous experimental results showed that a large number of lncRNAs were differentially expressed in both animal [31] and cell [32,33] models of PD.
This study also used microarray technology to assess whole-genome expression profiles of lncRNAs in Uyghur individuals with and without PD. Fifty differentially expressed lncRNAs were identified in these 2 groups, as were 97 mRNAs.
The relationship between lncRNA and PD is still unclear. Studies have found abnormal expression of lncRNAs in early PD [34,35], and antisense lncRNAs have been shown to regulate PD characteristics [36]. For example, NEAT1 lncRNA was found to inhibit the degradation of PINK1 protein, and interference with NEAT1 has been found to ameliorate damage to dopaminergic neurons [37]. Moreover, lncRNAs extracted from plasma exosomes were found to be differentially expressed. The results of bioinformatics analysis suggest that lnc-MKRN2–42 may be related to the occurrence and development of PD [13]. These studies also suggest that lncRNAs may be biomarkers for PD and may play important roles in the pathogenesis of PD. Further studies are needed to determine the role of lncRNAs in personalized neurology.
GO, KEGG enrichment, and pathway analyses are all important components of bioinformatics analysis. GO analysis of differentially expressed mRNAs of Uyghur PD patients and healthy controls found that the first few terms were closely related to biological processes, including cellular processes, single-organism processes, biological regulation, regulation of biological processes, and metabolic processes. In addition, the first few terms more closely related to the degree of cellular components included cells, parts of cells, organelles, and membranes. The first few terms more closely related to molecular functions included binding, catalytic activity, molecular transduction activity, and regulation of molecular function regulator. GO analysis initially addressed the biological information of genes with significantly different levels of expression in the PD and control groups at these 3 levels, providing direction for basic research on the pathogenesis of PD.
In this study, differentially expressed lncRNAs and mRNAs were selected by comparing their levels of expression in the Uyghur PD and healthy control groups. Pathway analysis revealed that the most enriched pathways corresponding to the dysregulation of lncRNAs related to PD were the inflammatory signaling pathway and its corresponding NF-kappa B signaling pathway. Inflammation plays an important role in the pathophysiology and etiology of neurodegenerative diseases [38,39]. Studies have suggested a possible connection between the loss of dopaminergic neurons and autoimmunity in PD [40,41]. Persistent inflammatory response is a major factor in the degeneration of dopaminergic neurons in PD [42,43]. Specific autoantibodies (AAbs) in PD may react with certain neuronal components involved in PD. Immunoregulatory therapy may have therapeutic significance for PD treatment in the future. Other dysregulated lncRNAs were related to the binding of chemokines to chemokine receptors and cytokine-cytokine receptor interactions, as well as their subsequent signaling pathways.
This study had several limitations, including its recruitment of participants from a single ethnic group in a single center in China. Therefore, it is unclear whether these differences in expression also occur in other sets of PD patients. Another limitation was the small sample size, indicating the need to validate these results in larger populations.
Conclusions
In conclusion, a microarray method was used to detect the expression of lncRNAs in the peripheral blood of Uyghur PD patients and healthy controls. The results provided biological information on lncRNA expression and the expression of corresponding mRNAs expression throughout the entire genome. The potential functional linkage of PD revealed that lncRNA expression was dysregulated and involved several biological and pathological processes. The abnormally expressed lncRNAs were associated with the regulation of inflammation and autoimmune diseases. The biological information and functional links provided by this study may provide clues to the pathogenesis and development of PD.
Figures
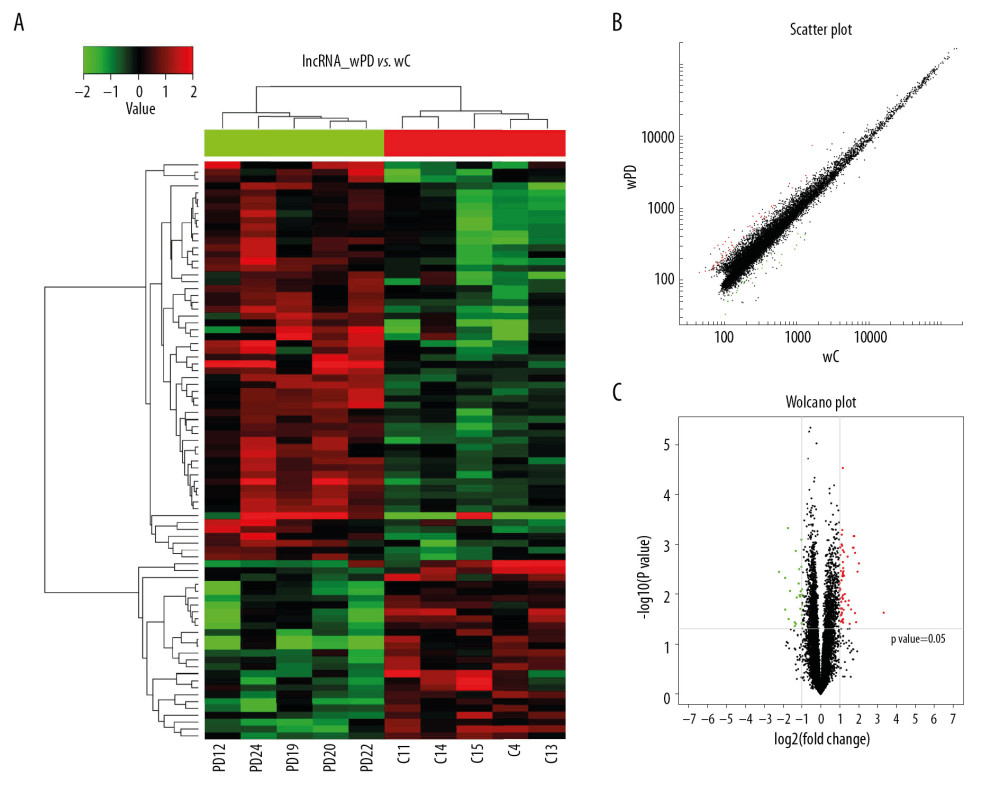 Figure 1. Hierarchical cluster analysis of lncRNAs differentially expressed in the PD and control groups. (A) Cluster analysis of lncRNAs. “Red” represents high relative expression, and “green” represents low relative expression in PD patients relative to controls. (B) Scatter plot of differential lncRNA expression. (C) Volcano plot of differential lncRNA expression.
Figure 1. Hierarchical cluster analysis of lncRNAs differentially expressed in the PD and control groups. (A) Cluster analysis of lncRNAs. “Red” represents high relative expression, and “green” represents low relative expression in PD patients relative to controls. (B) Scatter plot of differential lncRNA expression. (C) Volcano plot of differential lncRNA expression. 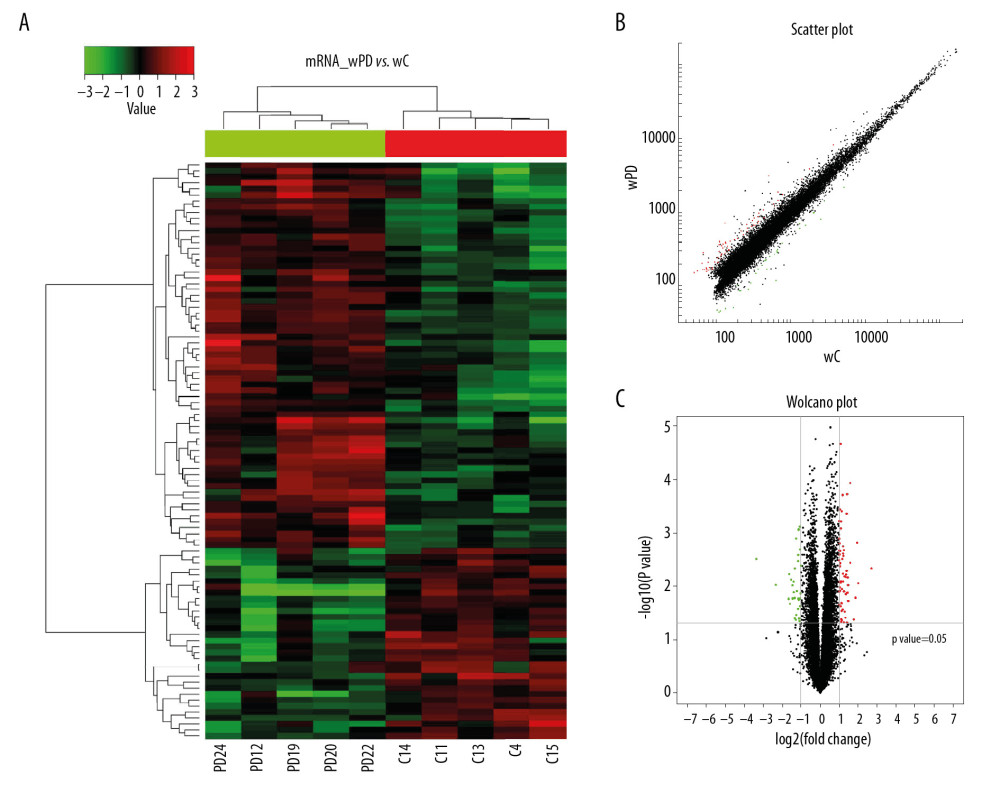 Figure 2. Hierarchical cluster analysis of mRNAs differentially expressed in the PD and control groups. (A) Cluster analysis of mRNAs. “Red” represents high relative expression, and “green” represents low relative expression in PD patients relative to controls. (B) Scatter plot of differential mRNA expression. (C) Volcano plot of differential mRNA expression.
Figure 2. Hierarchical cluster analysis of mRNAs differentially expressed in the PD and control groups. (A) Cluster analysis of mRNAs. “Red” represents high relative expression, and “green” represents low relative expression in PD patients relative to controls. (B) Scatter plot of differential mRNA expression. (C) Volcano plot of differential mRNA expression. 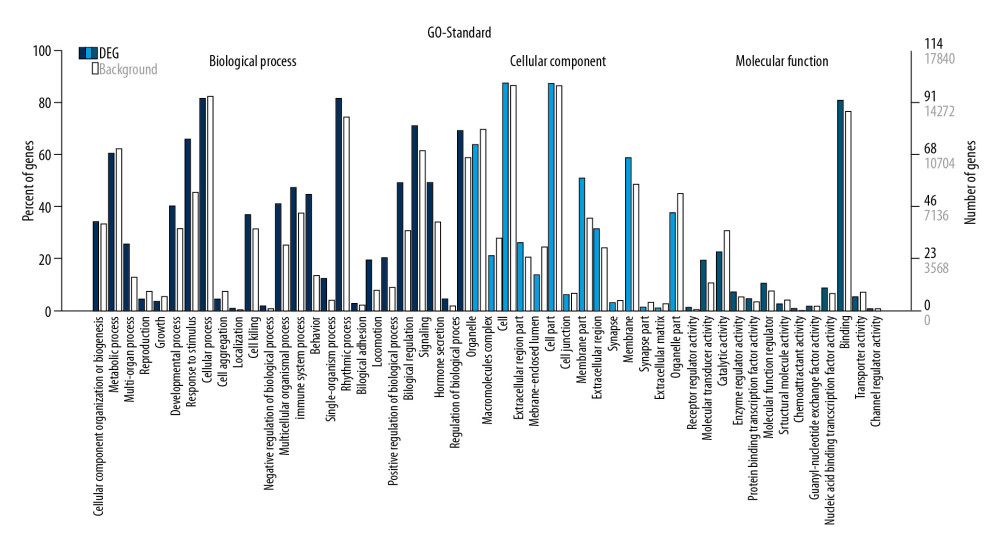 Figure 3. GO enrichment terms of differentially expressed lncRNAs in PD patients.
Figure 3. GO enrichment terms of differentially expressed lncRNAs in PD patients. 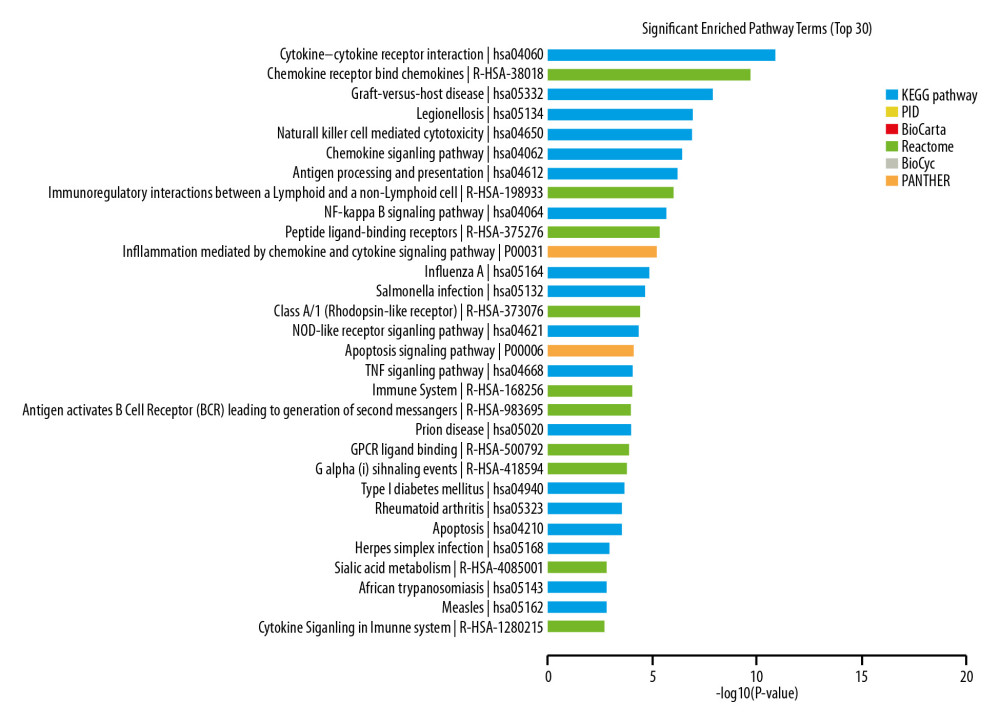 Figure 4. Pathway analysis of differentially expressed lncRNAs. Different colors represent different databases.
Figure 4. Pathway analysis of differentially expressed lncRNAs. Different colors represent different databases. 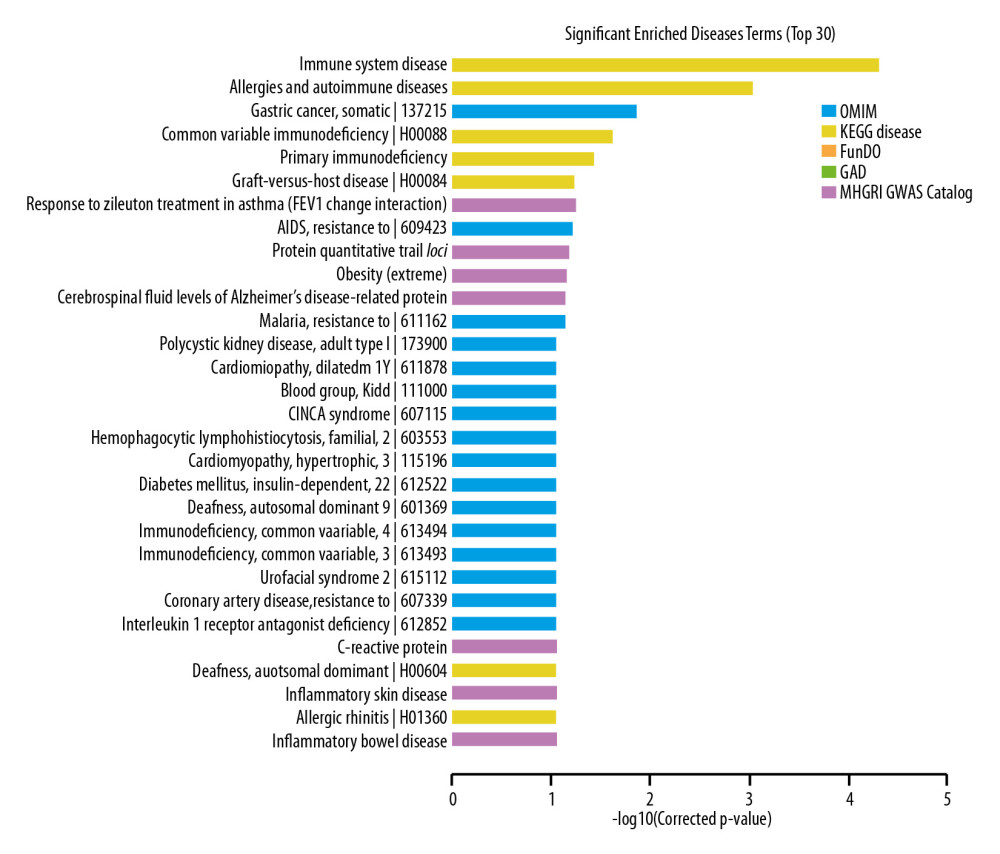 Figure 5. Disease analysis of differentially expressed lncRNAs.
Figure 5. Disease analysis of differentially expressed lncRNAs. 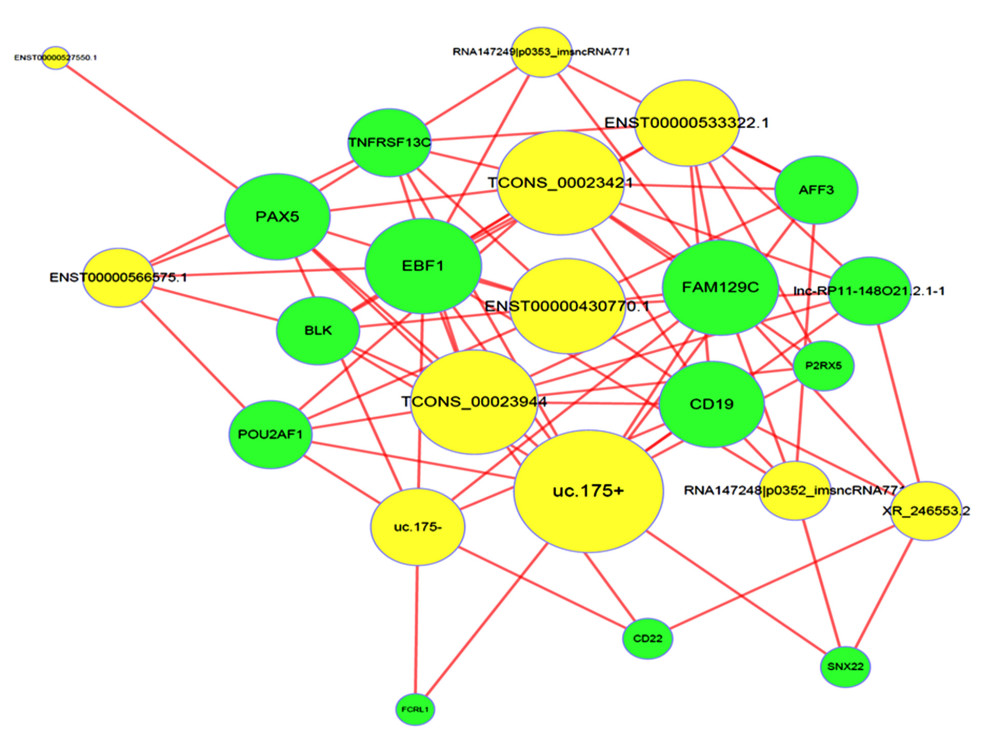 Figure 6. LncRNA-mRNA-network. Yellow dots indicate lncRNAs, and green nodes indicate target mRNAs.
Figure 6. LncRNA-mRNA-network. Yellow dots indicate lncRNAs, and green nodes indicate target mRNAs. Tables
Table 1. PCR primers used in the amplification of lncRNAs.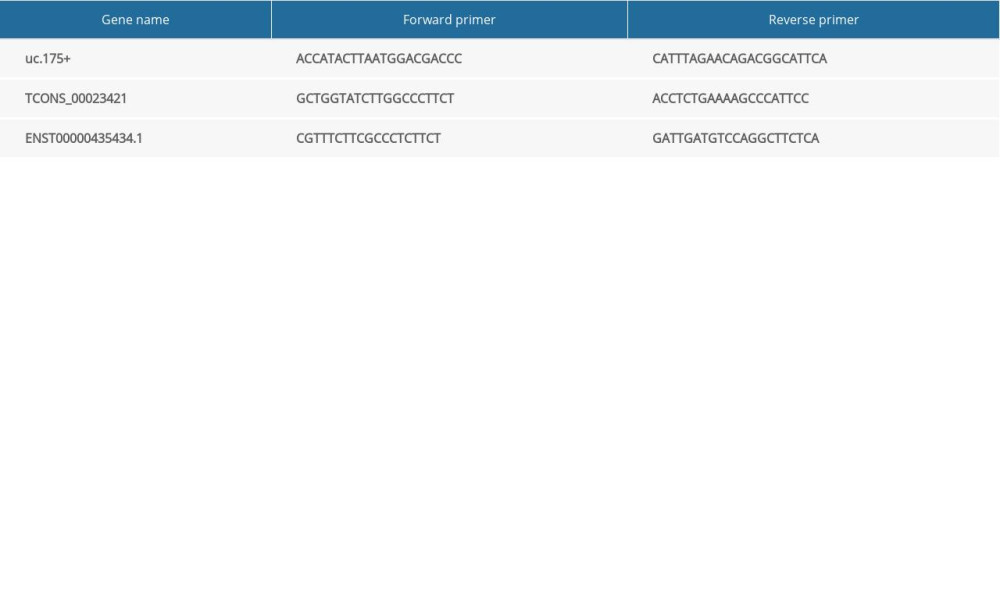 Table 2. Clinical characteristics of the included participants.
Table 2. Clinical characteristics of the included participants.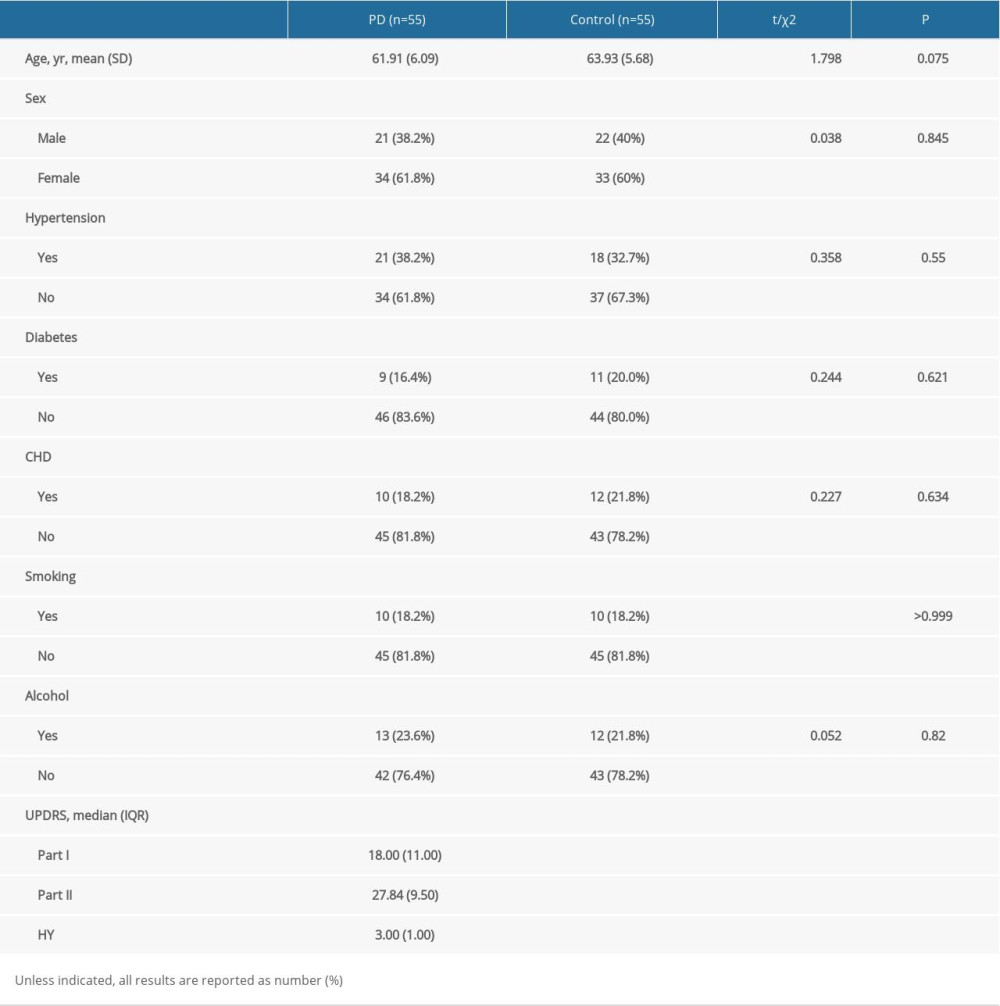 Table 3. LncRNAs differentially expressed in PD patients and controls.
Table 3. LncRNAs differentially expressed in PD patients and controls.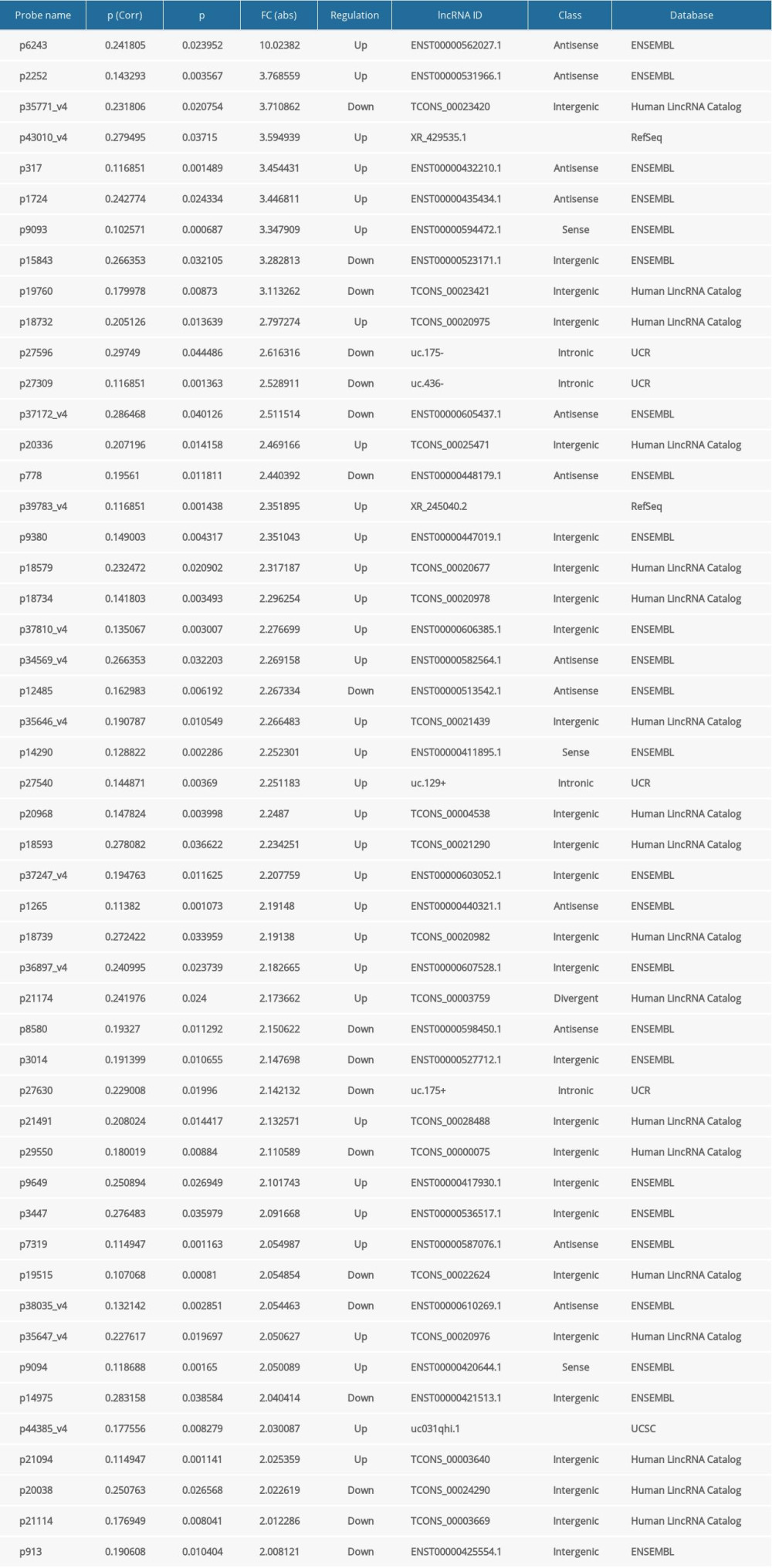 Table 4. The top 25 differentially expressed mRNAs in the PD and control groups.
Table 4. The top 25 differentially expressed mRNAs in the PD and control groups.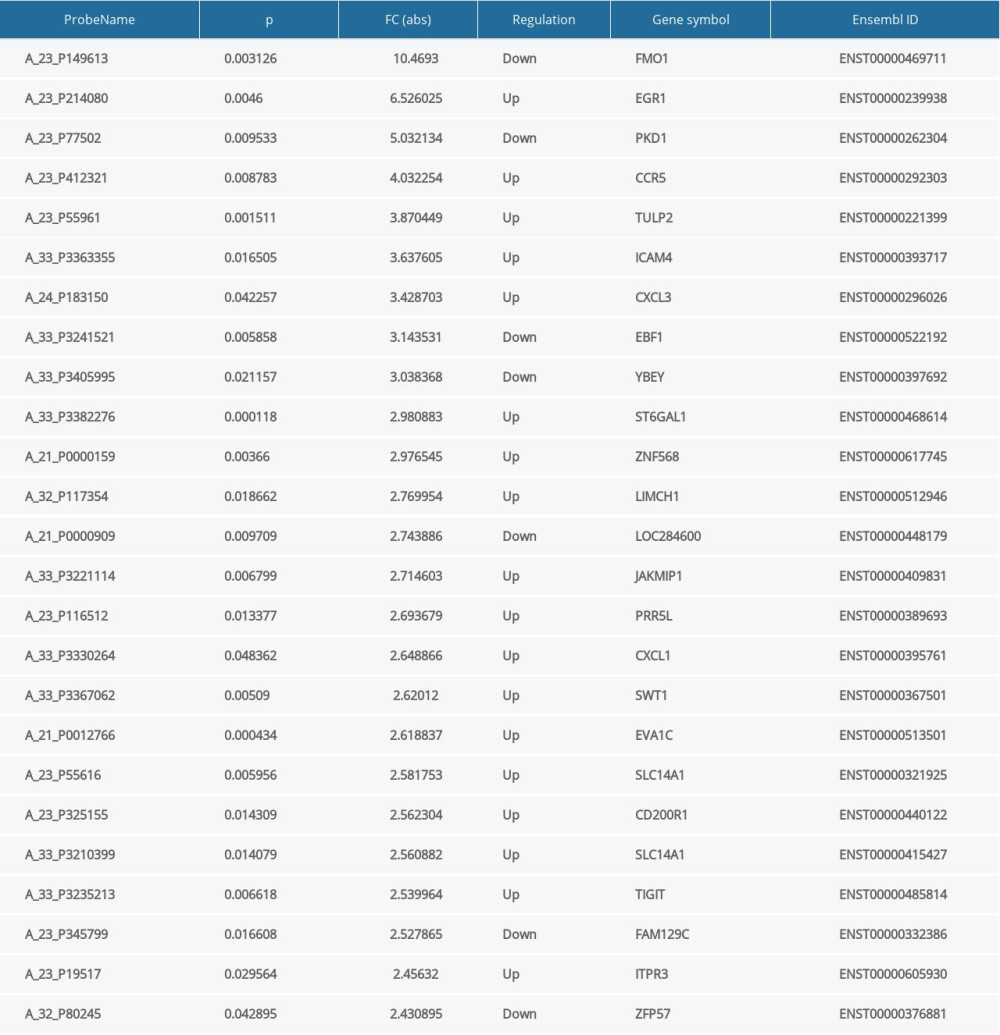 Table 5. Randomly selected lncRNAs.
Table 5. Randomly selected lncRNAs.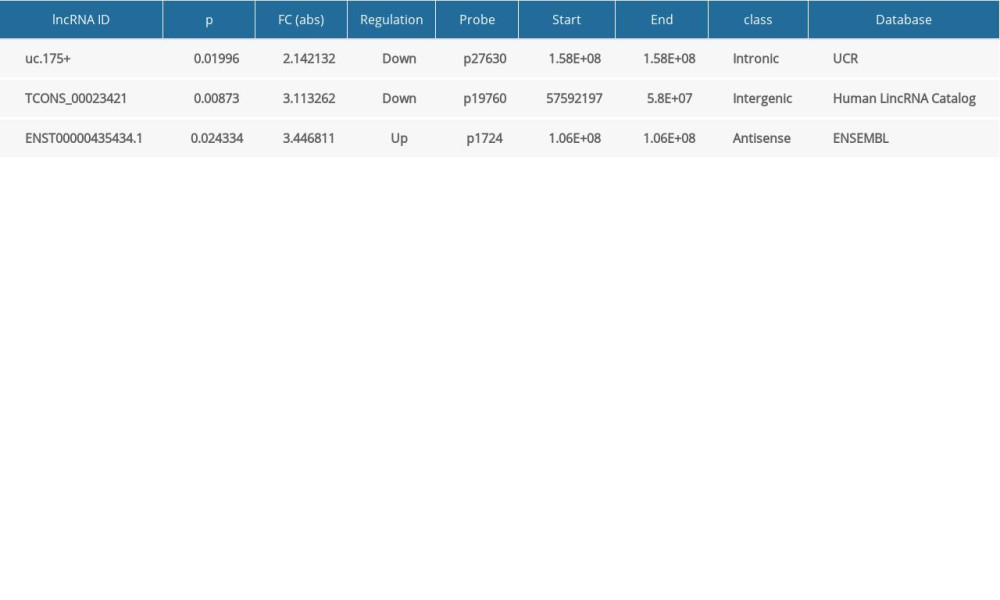
References
1. Kose Ozlece H, Findik Guvendi G, Huseyinoglu N, Cytological and cytometric analysis of oral mucosa in patients with Alzheimer’s and Parkinson’s disease: Neuropsychiatr Dis Treat, 2018; 14; 1901-6
2. Liu H, Liu J, Si L, GDF 15 promotes mitochondrial function and proliferation in neuronal HT22 cells: Cell Biochem, 2019; 120(6); 10530-47
3. Abbas MM, Xu Z, Tan LCS, Epidemiology of Parkinson’s disease-east versus west: Mov Disord Clin Pract, 2017; 5(1); 14-28
4. Ascherio A, Schwarzschild MA, The epidemiology of Parkinson’s disease: Risk factors and prevention: Lancet Neurol, 2016; 15(12); 1257-72
5. Brouwer M, Huss A, van der Mark M, Environmental exposure to pesticides and the risk of Parkinson’s disease in the Netherlands: Environ Int, 2017; 107; 100-10
6. Xu CF, Jiang LMolecular biological properties of lncRNA and their regulatory functions in the development of central nervous system: Journal of Shanghai Jiao Tong University (Medical Science), 2017; 37(2); 262-66
7. Ponting CP, Belgard TG, Transcribed dark matter: Meaning or myth?: Hum Mol Genet, 2010; 19(R2); R162-68
8. Li X, Wu Z, Fu X, Han W, Long noncoding RNAs: Insights from biological features and functions to diseases: Med Res Rev, 2013; 33(3); 517-53
9. Xue M, Zhuo Y, Shan B, MicroRNAs, long noncoding RNAs, and their functions in human disease: Methods Mol Biol, 2017; 1617; 1-25
10. Baker M, Long noncoding RNAs: The search for function: Nat Methods, 2011; 8(5); 379-83
11. Wu Y, Le W, Jankovic J, Preclinical biomarkers of Parkinson disease: Arch Neurol, 2011; 68(1); 22-30
12. Soreq L, Guffanti A, Salomonis N, Long non-coding RNA and alternative splicing modulations in Parkinson’s leukocytes identified by RNA sequencing: PloS Comput Biol, 2014; 10(3); e1003517
13. Wang Q, Han CL, Wang KL, Integrated analysis of exosomal lncRNA and mRNA expression profiles reveals the involvement of lnc-MKRN2-42: 1 in the pathogenesis of Parkinson’s disease: CNS Neurosci Ther, 2020; 26(5); 527-37
14. Zhai H, Li XM, Liu F, Expression pattern of genome-scale long noncoding RNA following acute myocardial infarction in Chinese Uyghur patients: Oncotarget, 2017; 8(19); 31449-64
15. Li G, Zhang Z, Xia H, Yang X, Analysis of Thr12Met and Ala1144Thr mutations of the ATP13A2 gene in Parkinson’s disease patients in Xinjiang Uygur and Han ethnic groups: Med Sci Monit, 2014; 20; 2177-82
16. Hughes AJ, Daniel SE, Kilford L, Lees AJ, Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: A clinico-pathological study of 100 cases: J Neurol Neurosurg Psychiatry, 1992; 55(3); 181-84
17. Sun J, Wu J, Expression profiling of long noncoding RNAs in neonatal and adult mouse testis: Data Brief, 2015; 4; 322-27
18. Gao W, Hillwig ML, Huang L, A functional genomics approach to tanshinone biosynthesis provides stereochemical insights: Org Lett, 2009; 11(22); 5170-73
19. Li H, Li H, Yue H, Comparison between smaller ruptured intracranial aneurysm and larger un-ruptured intracranial aneurysm: Gene expression profile analysis: Neurosurg Rev, 2017; 40(3); 419-25
20. Orom UA, Derrien T, Beringer M, Long noncoding RNAs with enhancer-like function in human cells: Cell, 2010; 143(1); 46-58
21. Kanehisa M, Sato Y, Kawashima M, KEGG as a reference resource for gene and protein annotation: Nucleic Acids Res, 2015; 44(D1); D457-62
22. Barabasi AL, Oltvai ZN, Network biology: Understanding the cell’s functional organization: Nat Rev Genet, 2004; 5(2); 101-13
23. Clark MB, Johnston RL, Inostroza-Ponta M, Genome-wide analysis of long noncoding RNA stability: Genome Res, 2012; 22(5); 885-98
24. Wang H, Zheng H, Azuaje F, Poisson-based self-organizing feature maps and hierarchical clustering for serial analysis of gene expression data: IEEE/ACM Trans Comput Biol Bioinform, 2007; 4(2); 163-75
25. Sato S, Li Y, Hattori N, Lysosomal defects in ATP13A2 and GBA associated familial Parkinson’s disease: J Neural Transm (Vienna), 2017; 124(11); 1395-400
26. Esteller M, Non-coding RNAs in human disease: Nat Rev Genet, 2011; 12(12); 861-74
27. Majidinia M, Mihanfar A, Rahbarghazi R, The roles of non-coding RNAs in Parkinson’s disease: Mol Biol Rep, 2016; 43(11); 1193-204
28. Salta E, De Strooper B, Non-coding RNAs with essential roles in neurodegenerative disorders: Lancet Neurol, 2012; 11(2); 189-200
29. Kraus TFJ, Haider M, Spanner J, Altered long noncoding RNA expression precedes the course of Parkinson’s disease-a preliminary report: Mol Neurobiol, 2017; 54(4); 2869-77
30. Ni Y, Huang H, Chen Y, Investigation of long non-coding RNA expression profiles in the substantia nigra of Parkinson’s disease: Cell Mol Neurobiol, 2017; 37(2); 329-38
31. Jiao F, Wang Q, Zhang P, Expression signatures of long non-coding RNA in the substantia nigra of pre-symptomatic mouse model of Parkinson’s disease: Behav Brain Res, 2017; 331; 123-30
32. Coupland KG, Kim WS, Halliday GM, Role of the long non-coding RNA MAPT-AS1 in regulation of microtubule associated protein tau (MAPT) expression in Parkinson’s disease: PLoS One, 2016; 11(6); e0157924
33. Zhang QS, Wang ZH, Zhang JL, Beta-asarone protects against MPTP-induced Parkinson’s disease via regulating long non-coding RNA MALAT1 and inhibiting α-synuclein protein expression: Biomed Pharmacother, 2016; 83; 153-59
34. Lin D, Liang Y, Jing X, Microarray analysis of an synthetic alpha-synuclein induced cellular model reveals the expression profile of long non-coding RNA in Parkinson’s disease: Brain Res, 2018; 1678; 384-96
35. Wang L, Yang H, Wang Q, Paraquat and MPTP induce alteration in the expression profile of long noncoding RNAs in the substantia nigra of mice: Role of the transcription factor Nrf2: Toxicol Lett, 2018; 291; 11-28
36. Carrieri C, Forrest AR, Santoro C: Front Cell Neurosci, 2015; 9; 114
37. Yan W, Chen ZY, Chen JQ, Chen HM, LncRNA NEAT1 promotes autophagy in MPTP-induced Parkinson’s disease through stabilizing PINK1 protein: Biochem Biophys Res Commun, 2018; 496(4); 1019-24
38. Holmans P, Moskvina V, Jones L, Sharma MInternational Parkinson’s Disease Genomics Consortium, A pathway-based analysis provides additional support for an immune-related genetic susceptibility to Parkinson’s disease: Hum Mol Genet, 2013; 22(5); 1039-49
39. von Euler Chelpin M, Vorup-Jensen T, Targets and mechanisms in prevention of Parkinson’s disease through immunomodulatory treatments: Scand J Immunol, 2017; 85(5); 321-30
40. De Virgilio A, Greco A, Fabbrini G, Corrigendum to “Parkinson’s disease: Autoimmunity and neuroinflammation” [Autoimmun Rev. 2016;15(10): 1005–1011]: Autoimmun Rev, 2016; 15(12); 1210
41. Gelders G, Baekelandt V, Van der Perren A, Linking neuroinflammation and neurodegeneration in Parkinson’s disease: J Immunol Res, 2018; 2018 4784268
42. Deleidi M, Gasser T, The role of inflammation in sporadic and familial Parkinson’s disease: Cell Mol Life Sci, 2013; 70(22); 4259-73
43. Kustrimovic N, Marino F, Cosentino M, Peripheral immunity, immunoaging and neuroinflammation in Parkinson’s disease: Curr Med Chem, 2019; 26(20); 3719-53
Figures
 Figure 1. Hierarchical cluster analysis of lncRNAs differentially expressed in the PD and control groups. (A) Cluster analysis of lncRNAs. “Red” represents high relative expression, and “green” represents low relative expression in PD patients relative to controls. (B) Scatter plot of differential lncRNA expression. (C) Volcano plot of differential lncRNA expression.
Figure 1. Hierarchical cluster analysis of lncRNAs differentially expressed in the PD and control groups. (A) Cluster analysis of lncRNAs. “Red” represents high relative expression, and “green” represents low relative expression in PD patients relative to controls. (B) Scatter plot of differential lncRNA expression. (C) Volcano plot of differential lncRNA expression. Figure 2. Hierarchical cluster analysis of mRNAs differentially expressed in the PD and control groups. (A) Cluster analysis of mRNAs. “Red” represents high relative expression, and “green” represents low relative expression in PD patients relative to controls. (B) Scatter plot of differential mRNA expression. (C) Volcano plot of differential mRNA expression.
Figure 2. Hierarchical cluster analysis of mRNAs differentially expressed in the PD and control groups. (A) Cluster analysis of mRNAs. “Red” represents high relative expression, and “green” represents low relative expression in PD patients relative to controls. (B) Scatter plot of differential mRNA expression. (C) Volcano plot of differential mRNA expression. Figure 3. GO enrichment terms of differentially expressed lncRNAs in PD patients.
Figure 3. GO enrichment terms of differentially expressed lncRNAs in PD patients. Figure 4. Pathway analysis of differentially expressed lncRNAs. Different colors represent different databases.
Figure 4. Pathway analysis of differentially expressed lncRNAs. Different colors represent different databases. Figure 5. Disease analysis of differentially expressed lncRNAs.
Figure 5. Disease analysis of differentially expressed lncRNAs. Figure 6. LncRNA-mRNA-network. Yellow dots indicate lncRNAs, and green nodes indicate target mRNAs.
Figure 6. LncRNA-mRNA-network. Yellow dots indicate lncRNAs, and green nodes indicate target mRNAs. Tables
 Table 1. PCR primers used in the amplification of lncRNAs.
Table 1. PCR primers used in the amplification of lncRNAs. Table 2. Clinical characteristics of the included participants.
Table 2. Clinical characteristics of the included participants. Table 3. LncRNAs differentially expressed in PD patients and controls.
Table 3. LncRNAs differentially expressed in PD patients and controls. Table 4. The top 25 differentially expressed mRNAs in the PD and control groups.
Table 4. The top 25 differentially expressed mRNAs in the PD and control groups. Table 5. Randomly selected lncRNAs.
Table 5. Randomly selected lncRNAs. Table 1. PCR primers used in the amplification of lncRNAs.
Table 1. PCR primers used in the amplification of lncRNAs. Table 2. Clinical characteristics of the included participants.
Table 2. Clinical characteristics of the included participants. Table 3. LncRNAs differentially expressed in PD patients and controls.
Table 3. LncRNAs differentially expressed in PD patients and controls. Table 4. The top 25 differentially expressed mRNAs in the PD and control groups.
Table 4. The top 25 differentially expressed mRNAs in the PD and control groups. Table 5. Randomly selected lncRNAs.
Table 5. Randomly selected lncRNAs. In Press
15 Apr 2024 : Laboratory Research
The Role of Copper-Induced M2 Macrophage Polarization in Protecting Cartilage Matrix in OsteoarthritisMed Sci Monit In Press; DOI: 10.12659/MSM.943738
07 Mar 2024 : Clinical Research
Knowledge of and Attitudes Toward Clinical Trials: A Questionnaire-Based Study of 179 Male Third- and Fourt...Med Sci Monit In Press; DOI: 10.12659/MSM.943468
08 Mar 2024 : Animal Research
Modification of Experimental Model of Necrotizing Enterocolitis (NEC) in Rat Pups by Single Exposure to Hyp...Med Sci Monit In Press; DOI: 10.12659/MSM.943443
18 Apr 2024 : Clinical Research
Comparative Analysis of Open and Closed Sphincterotomy for the Treatment of Chronic Anal Fissure: Safety an...Med Sci Monit In Press; DOI: 10.12659/MSM.944127
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952








