23 September 2023: Lab/In Vitro Research
Comparative Analysis of Sampling Methods for Assessing Bacterial Contamination on Hospital Partition Curtains: Moistened Swabs versus RODAC Agar Plates
Yaru Li1ACE, Leilei Li2CE, Feng Chen1BCD, Dong Wang1BF, Yaping Peng1BF, Wanqiu Wang1BF, Xiaxia Sun1BCF, Jianjun Deng1ACEG*, Juan Li1CFDOI: 10.12659/MSM.941086
Med Sci Monit 2023; 29:e941086
Abstract
BACKGROUND: Partition curtains are one of the main sources of nosocomial infection in the hospital environment. However, there are no unified standards for monitoring medical textiles across different countries or regions. This study aimed to investigate the accuracy of 2 different sampling methods – swabbing vs RODAC (replicate organism detection and counting) agar plate – in terms of detection of bacterial contamination, and their suitability as monitoring methods for partition curtains and other medical textiles.
MATERIAL AND METHODS: A total of 24 partition curtains were selected by stratified random sampling. The swabbing technique and RODAC agar plates were the chosen sampling methods. The number of colony-forming units was calculated and colony morphologies and strains on the plates were observed and identified after culturing.
RESULTS: A total of 192 samples were collected. Of them, 161 pathogenic strains were isolated via the swabbing technique and 309 pathogenic strains were isolated using the RODAC agar plates. The swabbing technique had a higher proportion for gram-positive bacteria (P=0.0004), while RODAC agar plates had a higher proportion for gram-negative bacteria (P=0.72). The detection of bacterial contamination rates using the swabbing technique was superior to that of the RODAC agar plate method (P<0.001).
CONCLUSIONS: The swabbing technique offers more advantages in terms of detection of bacterial contamination rates and gram-positive bacteria, while the RODAC agar plate is more sensitive for detection of gram-negative bacteria.
Keywords: Equipment Contamination, Environmental Monitoring, Textiles, Cross Infection, Humans, Agar, Bone Plates, Hospitals, Research Design
Background
Surfaces within the hospital environment have long been recognized as a potential repository of pathogens. Particularly, high-touch objects such as partition curtains and bed bars play an important role in the transmission of pathogens causing nosocomial infection [1]. Epidemiological evidence has shown that the risk of infection among newly admitted patients can be increased substantially if the surrounding environment is contaminated by pathogens such as
Cleaning and disinfection procedures are essential, since the concentration of pathogenic microorganisms in the hospital environment is higher than in normal social environments [5,6]. Evaluating the hospital environment in an effort to determine potential pathogens causing nosocomial infections can provide powerful evidence guiding the formulation of hospital environment monitoring criteria and guidelines for the continued prevention and control of nosocomial infection. In addition, environmental hygiene monitoring is a necessary means of evaluating the effects of routine cleaning and disinfection. ‘The Management of Multidrug-resistant Organisms in Medical Healthcare Setting’ (2006 Edition) issued by the US Healthcare Infection Control Practices Advisory Committee recommends continuously cleaning and disinfecting high-touch objects (level of evidence-based medicine: IB). Hospitals should adopt appropriate techniques for monitoring and inspecting the quality of cleaning and disinfection procedures in order to maintain a good hygienic environment in the hospital [7].
Partition curtains are one of the main sources of nosocomial infection in the hospital environment as they are high-touch objects. The results of Ohl et al have shown that partition curtains can be polluted to a hazardous extent within 1 week, with a contamination incidence of 92%; moreover, MRSA, VRE, and different types of gram-negative bacilli can be found on partition curtains [8]. Several studies have also reported that improper cleaning and disinfection of partition curtains can lead to outbreaks of nosocomial infection [9,10]. Pajamas and sheets were also identified as a potential source of nosocomial pathogens [11]. A report from Barzilai Hospital showed that bacterial colonization of sheets, including by MRSA, was found in 22 out of the 30 sheets examined [12]. In addition, healthcare worker attire can also become contaminated during contact with patients, with approximately 3–79% of healthcare worker gowns becoming contaminated with microorganisms, including
For many years, researchers and health practitioners have sought to develop and apply faster, more accurate methods for cleaning and disinfecting partition curtains in the hospital environment. The World Health Organization has designated several methods for scientific evaluation of environmental cleanliness, including swab culture, slide agar culture, direct observation, fluorescent labeling, and ATP biological fluorescence [2]. To date, there exist no unified standards for monitoring medical textiles across different countries or regions. In Germany, medical textile washing centers of medical institutions must possess a quality inspection and hygiene license issued by the Research Institute Hohenstein, authorized by the German Institute for Quality Assurance and Certification [14]. The Robert Koch Institute states that sampling using a RODAC agar plate is an acceptable means of monitoring pathogens on medical textiles [15,16]. In the UK, pathogens on textile products are counted using the membrane filtration method following recovery of the eluent of sterile textile products [17].
The swabbing technique is recommended in the Regulation for Washing and Disinfection Technique of Medical Textiles in Healthcare Facilities (WS/T508-2016) (transl.) in China [18]. Although corresponding policies have been put forward in different countries for streamlining cleaning and disinfection processes for medical textiles, and monitoring the biological load thereon, to date there exist no standardized techniques to guarantee appropriate levels of sensitivity and reliability [19].
In this study, we investigated the efficiency of 2 different sampling methods (swabbing vs RODAC agar plate) in terms of detection of bacterial contamination, and their suitability as monitoring methods for partition curtains and other medical textiles.
Material and Methods
SAMPLE COLLECTION:
The hospital in our study has 2 gynecological ward areas. During the time of investigation, a total of 103 beds in the 43 wards were in the gynecological ward areas. A total of 24 partition curtains were selected by stratified random sampling; 12 partition curtains were selected from each area between June 16, 2021 and July 14, 2021. The fold edge most often contacted by the hands of medical staff and patients was selected as the monitoring area. The swabbing technique and RODAC agar plates were the chosen sampling methods. The partition curtains were sampled 1 day, 1 week, 2 weeks, and 4 weeks after curtain cleaning.
SWABBING:
The swabbing technique was performed using chlorine and iodine-containing disinfectant neutralization medium produced by Wenzhou Kangtai Biotechnique Co., Ltd., China. The medium’s main components are beef powder, peptone, sodium chloride, and sodium thiosulfate. Referring to the Regulation for Washing and Disinfection Technique of Medical Textiles in Healthcare Facilities (WS/T508-20 16) (transl.), both sides of the bed curtain are exposed to the ward environment, and the 5cm × 5cm sterilization gauge board was placed in the selected sampling range. Then a swab wiping wet sampling solution (aseptic saline) was used to rub back and forth in the sterilization gauge board for a total of 5 times, The swab was rotated slowly in order to increase the contact area between the swab and partition curtains during this process. A total of 4 sampling points (none taken repeatedly) were collected from 2 areas on each side of the curtain. For each curtain, a total area of 100 cm2 was sampled. Sterilized scissors were used to cut off the part of the swab upper that the hand touched. The swabs with samples were put into a 10 mL sampling fluid tube for testing [18].
The tube filled with a swab with samples was eluted with sufficient oscillation. One mL of eluant was placed into a sterile plate. Then, in a temperature not exceeding 50°C, the nutrient agar was injected into the plate and the contents were shaken before being left to solidify. After culturing at 35°C for 48 h, the colonies were counted and the colony-forming units (CFUs) were calculated. In addition, colony morphologies and strains on the plates were observed and identified.
RODAC AGAR PLATE:
The RODAC agar plate technique employs the TSAWLP contact plate produced by Guangdong Huankai Microbial Co., Ltd., China. The contact plate was positioned on the selected area of the curtain, and the investigator used the finger to gently press the bottom of the plate center for 10 s. Two areas on each side of the curtain were sampled. Each plate was used to sample a 25 cm2 area. The total area sampled was 100 cm2. The plates with samples were put into a 35°C incubator for 48 h. Thereafter, the colonies were counted and the number of CFUs was calculated after culturing. Colony morphologies and strains on the plates were also observed and identified [20].
COLONY COUNTS AND PATHOGENIC BACTERIA IDENTIFICATION:
Colonies sampled using the swabbing technique were counted after 48 h of culturing, and the average colony count of each curtain was calculated. Colonies sampled using contact plates were counted after culturing in the RODAC agar plate, and the average colony count for each curtain was obtained by summing the 4 contact plates. According to the Regulation for Washing and Disinfection Technique of Medical Textiles in Healthcare Facilities (WS/T508-2016) (transl.), if the total number of bacterial colonies on the medical textiles is less than or equal to 200 CFU/100 cm2, and no pathogenic bacteria such as Staphylococcus aureus and coliform bacteria are detected, the curtain material is safe for use in the hospital environment [18]. The proportion of bacterial detection refers to the number of detected target bacteria, divided by all bacteria detected within the same period. The contamination rate refers to the number of curtains bearing bacterial levels exceeding the aforementioned thresholds, divided by the number of sampled curtains.
STATISTICAL ANALYSIS:
Measurement data were presented as the mean and standard deviation (χ±s). For normally distributed data, the t test was used, and the chi-square test was performed on enumeration data. The RM ANOVA test was used for repeatedly measured data to analyze the main effect, time effect, and interaction effect of processing factors [21]. The contrast test was performed when the interaction was found to be statistically significant (P<0.05). All analyses were performed using SAS software (SAS Institute), version 9.4.
Results
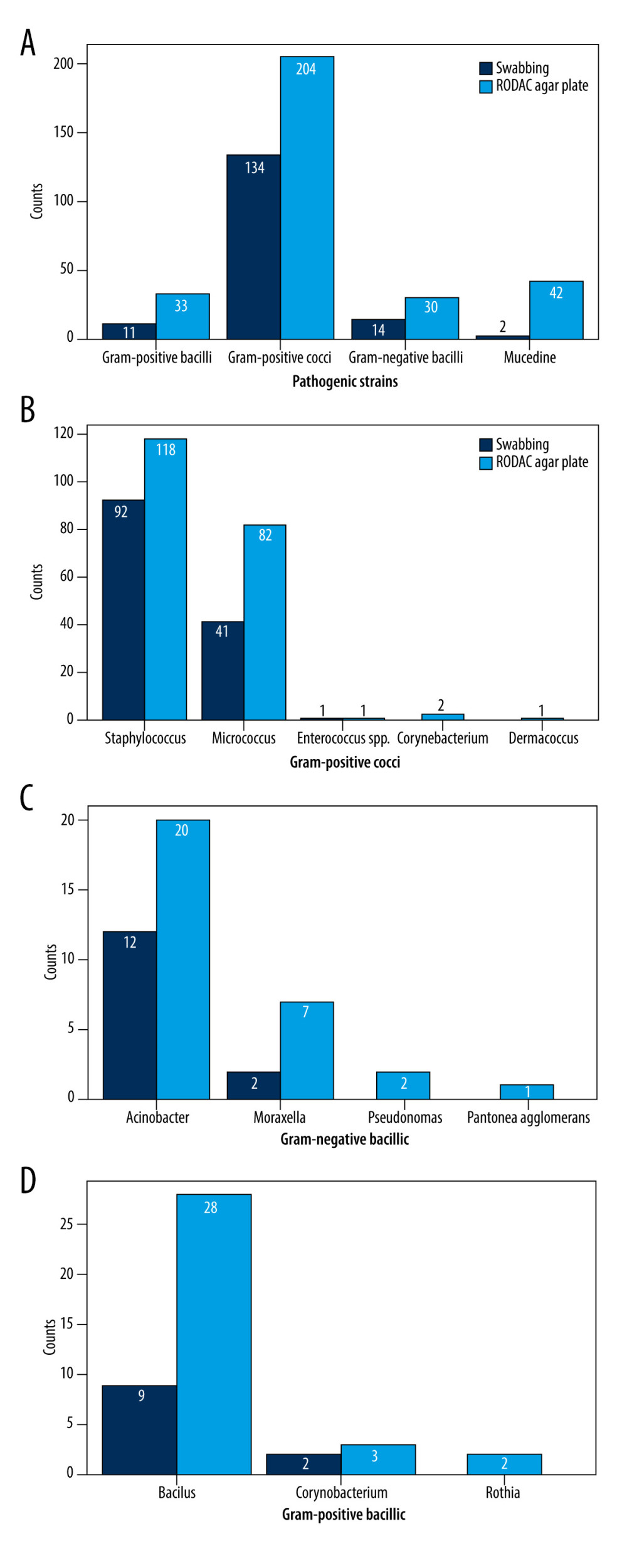
Of the 192 samples, 161 pathogenic strains, including 1
The swabbing technique detected a significant difference (
The bacterial contamination rates associated with the swabbing technique and RODAC agar plate were 30.21% and 0.02%, respectively. The detection of bacterial contamination rates using the swabbing technique was superior to that of the RODAC agar plate method, and a statistically significant difference (
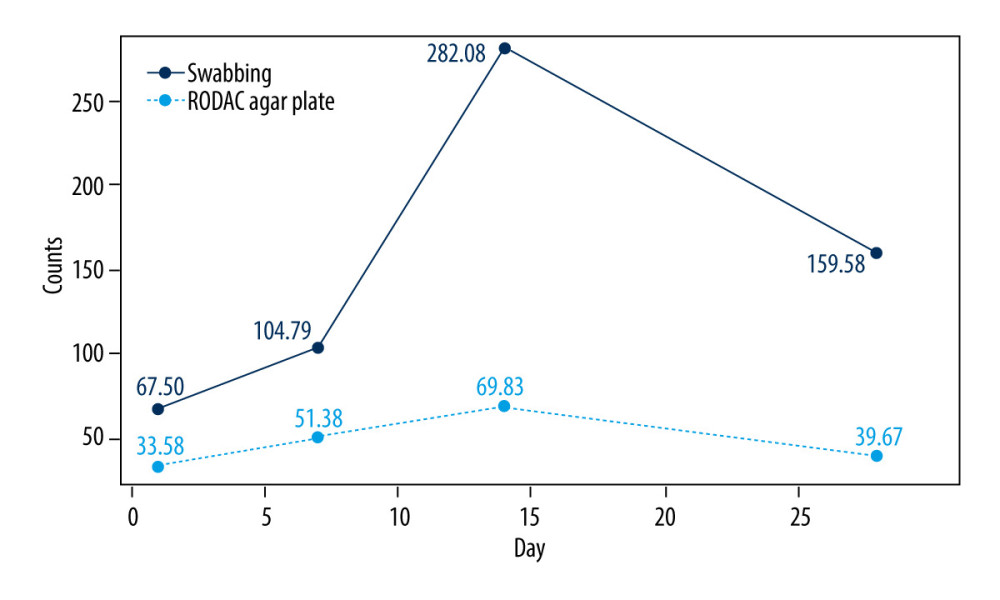
The results (Table 2) of RM ANOVA testing shows that the interactions between sampling method and time were statistically significant (
The number of bacteria on the septal curtain increased with time, reaching its highest on the 14th day, and then showed a downward trend (Figure 2). The results of the contrast test show (Table 4) that statistically significant differences were identified in first order (
Discussion
High-touch objects such as partition curtains, which are used widely in diagnosis and treatment areas, are sources of nosocomial infection. Appropriate techniques for monitoring such high-touch objects play a vital role in the prevention of cross-infection, thereby ensuring medical care quality and patient safety.
The results of our study are consistent with those of Lemmen et al, who reported that gram-positive bacteria are more common than gram-negative bacteria in hospital wards [20]. Gram-negative bacteria lose activity faster than gram-positive bacteria in dry conditions. Hence, with gram-positive bacteria there will be a wider contamination area on the surface of an object [22]. However, gram-negative bacteria such as
Obee et al found that the impact of the sampling method on the detection rate of the microorganisms on the environmental surface is greater than that of the culture medium used [25]. A study by Lemmen et al [20] showed that RODAC agar plates were more sensitive to gram-positive bacteria, with a detection rate of 69.5%, while swabbing technique was more sensitive to gram-negative bacteria, with a detection rate of 74.2%. However, in our study, the detection of gram-positive bacteria using the swabbing technique was superior to that of the RODAC agar plate method. We found no significant difference between the 2 methods in the detection of gram-negative bacteria, perhaps because of the low concentration of gram-negative bacteria on partition curtains. Rabuza et al compared the sampling efficiency of different sampling methods, and found the detection efficiency between the swabbing technique and the RODAC agar plate technique were different when the concentration of microorganisms on textile surfaces were varied. No microorganisms were found on the swabs at low concentrations [26].
It has been suggested that the number of CFUs detected on textile surfaces is lower than the actual concentration of microorganisms and there is a significant difference in accuracy among the various methods of textile sampling [27]. Rabuza et al [26] found that the swabbing technique gave the lowest results among the methods tested in their survey, but this disagrees with results of the present study. Our contrast test results and profile plot (Figure 2) show that there were lower counts with the RODAC agar plate method than with the swabbing technique. The results may be affected by the structure of the surface tested. Because the structure of a textile is a mechanical combination of warp and weft, a relatively small contact area is available, which leads to low CFU counts. Egington believed that the accuracy of RODAC agar plates depends on the surface tested. If surfaces are rough or wet, then the sampling is inaccurate or the resulting growth on the agar may be confluent [28]. In addition, the accuracy of the RODAC agar plate method may be affected by the microbial concentration on the surface tested. RODAC agar plates may be most accurate for surfaces with low microbial concentrations or for determining the existence of microorganisms [26].
Several studies suggested that 14 days is the minimum amount of time that curtains should hang before being cleaned [29,30], and this is also reflected in our study, as the numbers of bacteria on the partition curtains peaked at around the 14th day. Our study indicates that the number of bacteria in the curtain initially increased, then peaked and decreased over time, but this pattern contrasts with the research results of Shek et al [29]. Microorganism survival on fomites is influenced by intrinsic properties such as inoculum of the nosocomial pathogen, the type of surface material, and the type of suspension medium, and by extrinsic factors such as environmental temperature and humidity [3]. Moreover, there is a pervasive tradeoff between growth rate and competitive ability; therefore, it is predicted that the microbial community composition would change as the pollution rate of ward curtains changes [31]. In addition, because the first sampling would remove some of the bacteria from an area, leaving less bacteria for the subsequent sampling media, we chose to sample 2 adjacent sites where possible. This may have affected the results since the bacteria may not be present at each sample spot.
Conclusions
Overall, gram-positive bacteria are more common in hospital wards than in most other social environments. To evaluate pathogenic contaminants more accurately, the results of our study suggest that the 2 sampling methods should be combined. Our results show that the swabbing technique offers more advantages in terms of estimation of bacterial contamination rates, while the RODAC agar plate is more sensitive to the detection of different strains under limited conditions. Different sampling methods should be considered according to monitoring purposes.
Tables
Table 1. Accuracy of swabbing technique vs RODAC agar plate in detecting gram-positive and gram-negative bacteria. Table 2. Analysis of RM ANOVA test results of the colony counts of swabbing technique and RODAC agar plate.
Table 2. Analysis of RM ANOVA test results of the colony counts of swabbing technique and RODAC agar plate.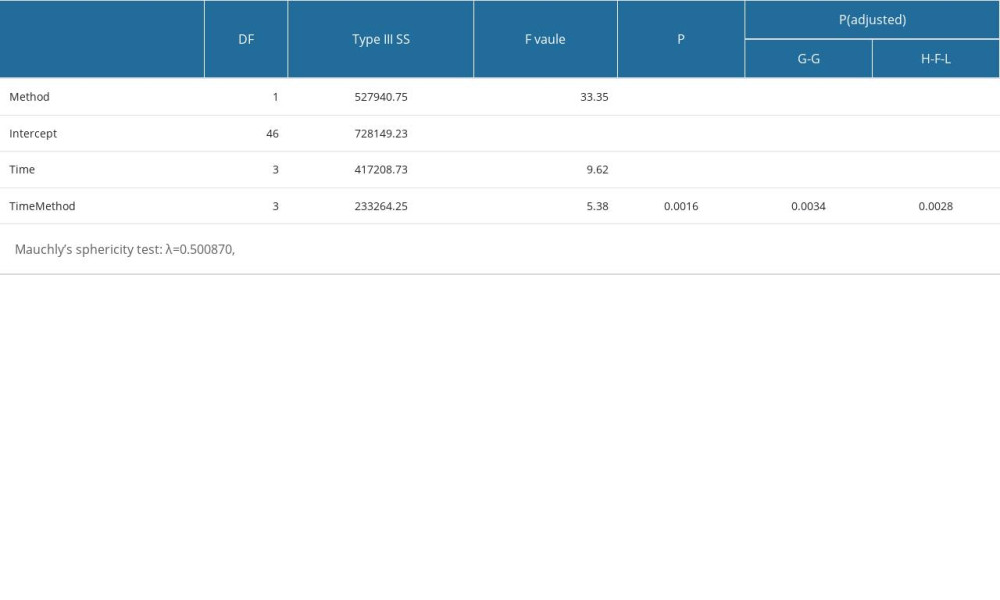 Table 3. The colony counts of swabbing technique and RODAC agar plate at different time points.
Table 3. The colony counts of swabbing technique and RODAC agar plate at different time points.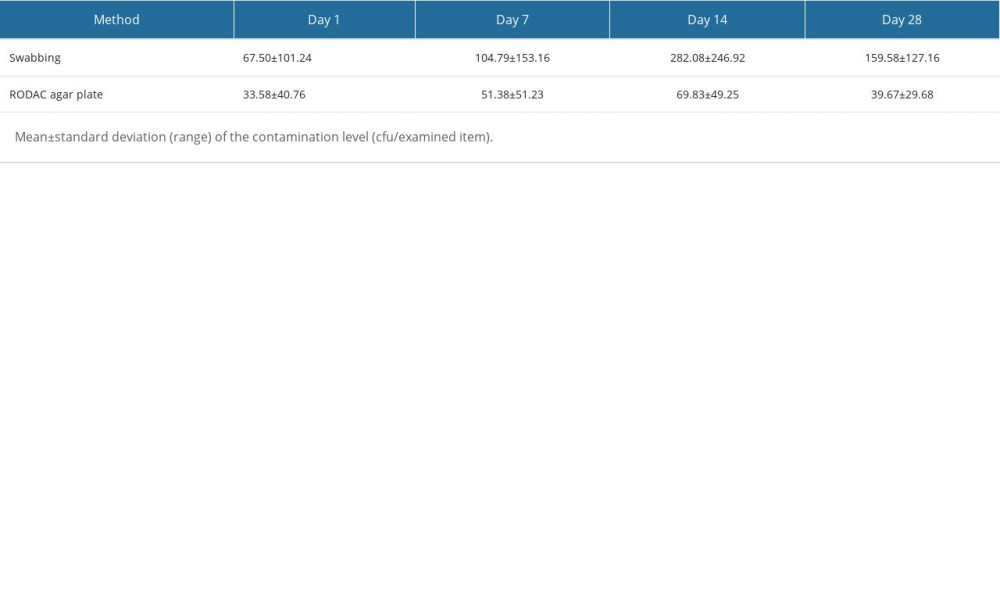 Table 4. Analysis of contrast test results of the colony counts of swabbing technique and RODAC agar plate.
Table 4. Analysis of contrast test results of the colony counts of swabbing technique and RODAC agar plate.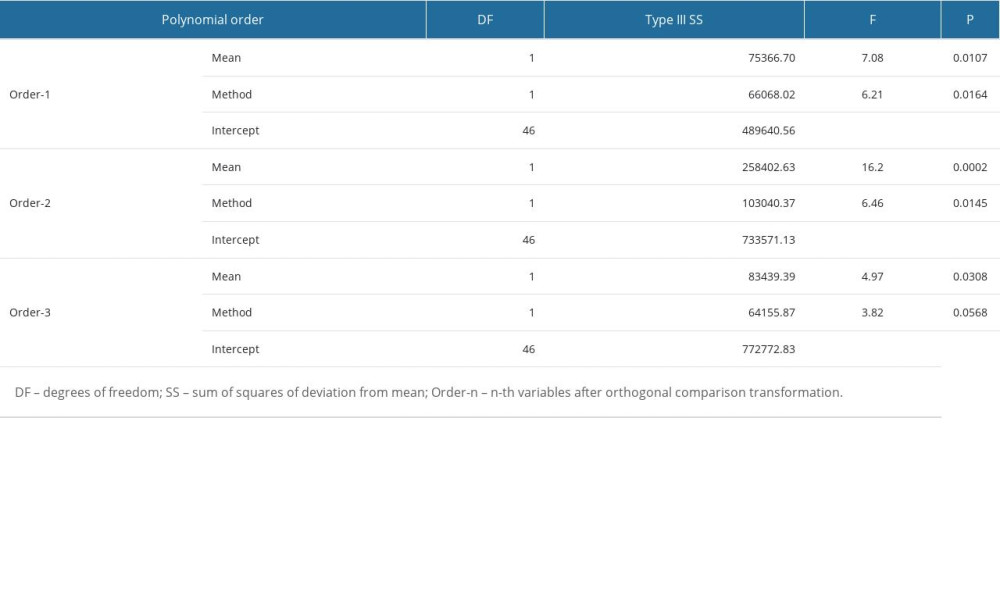
References
1. Cheng VC, Chau PH, Lee WM, Hand-touch contact assessment of high-touch and mutual-touch surfaces among healthcare workers, patients, and visitors: J Hosp Infect, 2015; 90(3); 220-25
2. Carling PC, Bartley JM, Evaluating hygienic cleaning in health care settings: What you do not know can harm your patients: Am J Infect Control, 2010; 38; S41-S50
3. Kramer A, Schwebke I, Kampf G, How long do nosocomial pathogens persist on inanimate surfaces? A systematic review: BMC Infect Dis, 2006; 6; 130
4. Dancer SJ, Kramer A, Four steps to clean hospitals: LOOK, PLAN, CLEAN and DRY: J Hosp Infect, 2019; 103(1); e1-e8
5. Datta R, Platt R, Yokoe DS, Huang SS, Environmental cleaning intervention and risk of acquiring multidrug-resistant organisms from prior room occupants: Arch Intern Med, 2011; 171; 491-94
6. Rutala WA, Weber DJ, Best practices for disinfection of noncritical environmental surfaces and equipment in health care facilities: A bundle approach: Am J Infect Control, 2019; 47S; A96-A105
7. Grota PG, Perioperative management of multidrug-resistant organisms in health care settings: AORN J, 2007; 86; 361-72
8. Ohl M, Schweizer M, Graham M, Hospital privacy curtains are frequently and rapidly contaminated with potentially pathogenic bacteria: Am J Infect Control, 2012; 40; 904-6
9. Mahida N, Beal A, Trigg D, Outbreak of invasive group A streptococcus infection: Contaminated patient curtains and cross-infection on an ear, nose and throat ward: J Hosp Infect, 2014; 87; 141-44
10. Das I, Lambert P, Hill D, Carbapenem-resistant Acinetobacter and role of curtains in an outbreak in intensive care units: J Hosp Infect, 2002; 50; 11
11. Malnick S, Bardenstein R, Huszar M, Pajamas and sheets as a potential source of nosocomial pathogens: J Hosp Infect, 2008; 70; 89-92
12. Gabbay J, Borkow G, Mishal J, Copper oxide impregnated textiles with potent biocidal activities: Journal of Industrial Textiles, 2006; 35; 323-35
13. Haun N, Hooper-Lane C, Safdar N, Healthcare personnel attire and devices as fomites: A systematic review: Infect Control Hosp Epidemiol, 2016; 37; 1367-73
14. Deutsches Institut für Gütezicherung und Kennzeichnung e.V Sachegemëße Waschepflege, Gütezicherung RAL-GZ 992: Proper Linen Care, Quality Assurance RAL-GZ 992, 2001, Sankt Avgustin, RAL
15. Heintz M, Bohnen J, Hygiene in commercial laundries: Hygiene & Medizin, 2011; 36; 292-99
16. Fijan S, Sostar-Turk S, Cencic A, Implementing hygiene monitoring systems in hospital laundries in order to reduce microbial contamination of hospital textiles: J Hosp Infect, 2005; 61; 30-38
17. : Textiles-Laundry processed textiles-Biocontamination control system. UNE-EN 14065-2017, 2017
18. : WS/T 508-2016, Regulation for washing and disinfection technique of medical textiles in healthcare facilities, 2016 Available at http://www.nhc.gov.cn/ewebeditor/uploadfile/2017/01/20170105092028826.pdf
19. Owen L, Laird K, The role of textiles as fomites in the healthcare environment: A review of the infection control risk: Peer J, 2020; 8; e9790
20. Lemmen SW, Häfner H, Zolldann D, Comparison of two sampling methods for the detection of gram-positive and gram-negative bacteria in the environment: moistened swabs versus Rodac plates: Int J Hyg Environ Health, 2001; 203; 245-48
21. Songlin Y, Huiyun X: Analysis of repeated measures data with SAS procedures, 2004, Beijing, Science Press
22. Hirai Y, Survival of bacteria under dry conditions; from a viewpoint of nosocomial infection: J Hosp Infect, 1991; 19; 191-200
23. Gaynes R, Edwards JRNational Nosocomial Infections Surveillance System, Overview of nosocomial infections caused by gram-negative bacilli: Clin Infect Dis, 2005; 41; 848-54
24. : Healthcare-Associated Infections (HAIs): Gram-negative bacteria infections in healthcare settings January, 2011, Centers for Disease Control and Prevention Available at https://www.cdc.gov/hai/organisms/gram-negative-bacteria.html
25. Obee P, Griffith CJ, Cooper RA, Bennion NE, An evaluation of different methods for the recovery of methicillin-resistant Staphylococcus aureus from environmental surfaces: J Hosp Infect, 2007; 65; 35-41
26. Rabuza U, Šostar–zTurk S, Fijan S, Efficiency of four sampling methods used to detect two common nosocomial pathogens on textiles: Textile Research Journal, 2012; 82; 2099-105
27. Rozman U, Fijan S, Turk SŠ, Mlakar V, Real-time polymerase chain reaction for quantitative assessment of common pathogens associated with healthcare-acquired infections on hospital textiles: Textile Research Journal, 2013; 83; 2032-41
28. Eginton PJ, Gibson H, Holah J, Handley PS, Gilbert P, Quantification of the ease of removal of bacteria from surfaces: J Ind Microbiol, 1995; 15; 305-10
29. Shek K, Patidar R, Kohja Z, Rate of contamination of hospital privacy curtains in a burns/plastic ward: A longitudinal study: Am J Infect Control, 2018; 46; 1019-21
30. Kurashige EJ, Oie S, Furukawa H: Braz J Microbiol, 2016; 47; 703-5
31. Abreu CI, Friedman J, Andersen Woltz VL, Gore J, Mortality causes universal changes in microbial community composition: Nat Commun, 2019; 10; 2120
Figures
Tables
 Table 1. Accuracy of swabbing technique vs RODAC agar plate in detecting gram-positive and gram-negative bacteria.
Table 1. Accuracy of swabbing technique vs RODAC agar plate in detecting gram-positive and gram-negative bacteria. Table 2. Analysis of RM ANOVA test results of the colony counts of swabbing technique and RODAC agar plate.
Table 2. Analysis of RM ANOVA test results of the colony counts of swabbing technique and RODAC agar plate. Table 3. The colony counts of swabbing technique and RODAC agar plate at different time points.
Table 3. The colony counts of swabbing technique and RODAC agar plate at different time points. Table 4. Analysis of contrast test results of the colony counts of swabbing technique and RODAC agar plate.
Table 4. Analysis of contrast test results of the colony counts of swabbing technique and RODAC agar plate. Table 1. Accuracy of swabbing technique vs RODAC agar plate in detecting gram-positive and gram-negative bacteria.
Table 1. Accuracy of swabbing technique vs RODAC agar plate in detecting gram-positive and gram-negative bacteria. Table 2. Analysis of RM ANOVA test results of the colony counts of swabbing technique and RODAC agar plate.
Table 2. Analysis of RM ANOVA test results of the colony counts of swabbing technique and RODAC agar plate. Table 3. The colony counts of swabbing technique and RODAC agar plate at different time points.
Table 3. The colony counts of swabbing technique and RODAC agar plate at different time points. Table 4. Analysis of contrast test results of the colony counts of swabbing technique and RODAC agar plate.
Table 4. Analysis of contrast test results of the colony counts of swabbing technique and RODAC agar plate. In Press
11 Mar 2024 : Clinical Research
Enhancement of Frozen-Thawed Human Sperm Quality with Zinc as a Cryoprotective AdditiveMed Sci Monit In Press; DOI: 10.12659/MSM.942946
12 Mar 2024 : Database Analysis
Risk Factors of Age-Related Macular Degeneration in a Population-Based Study: Results from SHIP-TREND-1 (St...Med Sci Monit In Press; DOI: 10.12659/MSM.943140
12 Mar 2024 : Clinical Research
Preoperative Blood Transfusion Requirements for Hemorrhoidal Severe Anemia: A Retrospective Study of 128 Pa...Med Sci Monit In Press; DOI: 10.12659/MSM.943126
12 Mar 2024 : Clinical Research
Tissue Inhibitors of Metalloproteinase 1 (TIMP-1) and 3 (TIMP-3) as New Markers of Acute Kidney Injury Afte...Med Sci Monit In Press; DOI: 10.12659/MSM.943500
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952