28 June 2020: Molecular Biology
Key Genes Associated with Non-Alcoholic Fatty Liver Disease and Acute Myocardial Infarction
Weiran Dai1DEF, Yue Sun2ABC, Zhiyuan Jiang3FG, Kuan Du2BD, Ning Xia2A*, Guoqiang Zhong1EGDOI: 10.12659/MSM.922492
Med Sci Monit 2020; 26:e922492
Abstract
BACKGROUND: With the increasing research on non-alcoholic fatty liver (NAFLD) and acute myocardial infarction (AMI), many evidences show a tight correlation between NAFLD and AMI, however the underlying pathophysiology is still not clear. This study was to identify the potential hub genes and pathways related to these two diseases via bioinformatic method.
MATERIAL AND METHODS: Gene Expression Omnibus (GEO) dataset GSE63067 of NAFLD patients and normal controls was downloaded from GEO database. The GSE60993 and GSE66360 datasets for AMI patients and healthy controls were also obtained. Differential expression genes (DEGs) of NAFLD and AMI datasets, accompanied with common genes between them were achieved. Further GO and KEGG enrichment analysis for common genes were performed.
RESULTS: To obtain the pathogenesis associated with both NAFLD and AMI, protein-protein interaction (PPI) network was constructed and the top ten hub genes (TLR2, LILRB2, CXCL1, FPR1, TLR4, TYROBP, MMP9, FCER1G, CLEC4D and CCR2) were selected with
CONCLUSIONS: The results of this study suggesting some novel genes may play an important role in the occurrence and progression NAFLD and AMI. But more experimental researches and clinical trials need to verify.
Keywords: Biological Markers, Liver Diseases, Cluster Analysis, Databases, Genetic, gene ontology, Non-alcoholic fatty liver disease, Protein Interaction Maps
Background
Non-alcoholic fatty liver disease (NAFLD) is a metabolic syndrome characterized by multiple definite causes, including cardiovascular and cerebrovascular diseases, DM, dietary rhythm, and environmental and genetic factors [1]. It was once considered as a benign compensatory disease; however, with the deepening of understanding of the disease, it is now known that it can progress from simple steatohepatitis to non-alcoholic steatohepatitis (NASH), and even cirrhosis and liver cancer [2]. Although its pathogenesis is unclear, inflammatory factors play an important role [3]. With the high prevalence of NAFLD, the number of patients and financial expenditure are increasing steadily, which imposes a heavy economic burden on society [4,5].
Acute myocardial infarction (AMI) is one of the main causes of death in developed countries [6]. Although therapeutic techniques have rapidly progressed, the morbidity and mortality rates remain at nearly 5% [7]. To date, the pathogenesis of AMI has not yet been fully studied; however, it is clear that inflammatory factors are related to the injury of cardiomyocytes and AMI [8].
In recent studies, AMI has been proved to be the most common cause of death among NAFLD patients [9,10]. A large-cohort study was performed on NAFLD patients in Korea, showing that AMI was more likely to occur in NAFLD patients compared with those without NAFLD after adjusting for potential risk factors of AMI and NAFLD [11]. Also, a meta-analysis including over than 34 000 participants revealed the odds ratio was 1.64 (95% confidence interval 1.26–2.13) for combined fatal and non-fatal AMI [12]. Many NAFLD patients have abnormally high glucose and lipid levels, as well as high BMI, which are both risk factors for AMI [13]. Although research on the relationship between NAFLD and AMI has progressed steadily, the related genetics research is still limited and needs further exploration.
In the present study, 1 gene expression profile for NAFLD and 2 profiles for AMI were downloaded from the Gene Expression Omnibus (GEO) database, which is an open-access database that provides genetics information and can be used to identify new targets of disease by bioinformatics analysis [14]. Differentially expressed genes (DEGs) between disease samples and normal controls in NAFLD and AMI were found, and the common expressed genes of these 2 diseases were obtained as well. Cluster analysis was conducted to search for hub genes with multiple functions, including Gene Ontology (GO) term enrichment, Kyoto Encyclopedia of Genes and Genomes (KEGG), and protein-protein interaction (PPI) network analyses. Consequently, the identified hub genes may become a novel research focus, and the obtained molecular mechanisms and signal pathways may also help to explain the relationships between NAFLD and AMI.
Material and Methods
MICROARRAY DATA COLLECTION:
The microarray datasets of NAFLD, AMI, and normal controls were downloaded from the Gene Expression Omnibus (GEO) database (
IDENTIFICATION OF DIFFERENTIALLY EXPRESSED GENES (DEGS) IN NAFLD AND AMI:
DEGs between NAFLD and normal controls, AMI patients, and corresponding controls were identified using the limma R package, which is an efficient analysis method in bioinformatics [15]. The selected criteria in NAFLD datasets were set as P-value <0.05 and |log2FC| >0.5. In AMI section, the cut-off values were adjusted-P<0.05 and |log2FC| >1. After we utilized these screening conditions, 2 sets of DEGs were identified, then we put these DEGs that came from the 2 diseases into an online analysis tool Venn (http://bioinformatics.psb.ugent.be/webtools/Venn/) to obtain their intersection genes. These intersecting common genes were used for subsequent analysis.
FUNCTIONAL ENRICHMENT ANALYSES FOR COMMON DEGS:
The Gene Ontology (GO) [16] classification, which contains molecular functions, biological processes, and cellular component, and the Kyoto Encyclopedia of Genes and Genomes (KEGG) [17] pathway enrichment analyses were performed on the intersecting common DEGs using the R package. P-values <0.05 were defined as statistically significant.
CONSTRUCTION OF PROTEIN–PROTEIN INTERACTION (PPI) NETWORK AND IDENTIFICATION OF HUB GENES:
To further explore the interaction among the common genes obtained above, we used the Search Tool for the Retrieval of Interacting Genes (STRING) 11.0 (http://string-db.org/) [18] to construct a PPI network. The minimum required interaction score was considered high confidence (0.700) as the criteria of statistical significance. Cytoscape 3.6.1 (https://cytoscape.org) [19] was utilized to present the results. In the network outcome, the nodes represented the proteins, while the lines represented the interactions between proteins [20]. After we identified the hub genes among these common genes, we installed the cytoHubba plugin (http://hub.iis.sinica.edu.tw/cytohubba/), which can be download for free via Cytoscape software [21].
STATISTICAL ANALYSIS:
Using SPSS 22.0 (SPSS, Inc, Chicago, IL, USA), we constructed receiver operating characteristic (ROC) curves and calculated the area under the curve (AUC) of the hub genes to compare the AUC as the index of the model. These results showed the diagnostic efficiency of genes. P-values <0.05 were regarded as statistically significant.
Results
DIFFERENTIAL EXPRESSION ANALYSIS OF DEGS IN NAFLD AND AMI:
In the NAFLD dataset GSE63067, 823 DEGs were filtered when we compared the 11 NAFLD samples with 7 normal controls. Two AMI-related datasets (GSE60993 and GSE66360) were enrolled in the study, the merged dataset contained 56 AMI patient samples and 57 of their control samples. Heatmaps showing the gene expression profiles of NAFLD and AMI are presented in Figures 1 and 2, after homogenization, there were 823 and 200 DEGs achieved with adjusted-P<0.05, respectively. Using the Venn Diagram online tool, 78 intersecting common genes of 2 diseases were obtained and are shown in Figure 3. The details of these common genes are also shown in the Supplementary Table 1. The information on the whole study process is presented in Figure 4.
GO AND KEGG ENRICHMENT PATHWAY ANALYSIS:
Functional enrichment and KEGG pathway analyses of 78 common genes were performed in NAFLD and AMI at the threshold of P-value <0.05. Changes in GO biological processes (BP) mainly included inflammatory reactions (e.g., inflammatory response, innate immune response, immune response, and neutrophil chemotaxis) and signal transduction, shown in Figure 5. In Figure 6, changes in cellular component (CC) were notably focused on enrichment of cell outer membranes, such as plasma membrane, integral component of plasma membrane, extracellular space, cell surface, and anchored component of membranes. Moreover, in the molecular function (MF) section (Figure 7), changes were significant in receptor activity (RAGE receptor binding and signaling pattern recognition receptor activity), binding-related function (carbohydrate binding and IgG binding), and pantetheine hydrolase activity. In particular, changes in the KEGG pathway were mostly enriched in several immune system diseases (leishmaniasis, tuberculosis, malaria, and rheumatoid arthritis), immune-related pathways (IL-17 signaling pathway and NF-kappa B signaling pathway), and transcriptional mis-regulation in cancer, which are illustrated by a dot plot in Figure 8.
PPI NETWORK ANALYSIS AND HUB GENE SELECTION:
To distinguish the hub genes from the common genes, a PPI network was constructed. As seen in Figure 9, toll-like receptor 2 (TLR2), leukocyte immunoglobulin-like receptor subfamily B2 (LILRB2), C-X-C motif chemokine ligand 1 (CXCL1), formyl peptide receptor 1 (FPR1), toll-like receptor 4 (TLR4), tyrosine kinase binding protein (TYROBP), matrix metalloproteinase 9 (MMP9), Fc-receptor common gamma chain (FCER1G), C-type lectin domain family 4 member D (CLEC4D), and chemokine receptor 2(CCR2) proteins interact with other proteins by >5, which was the central node of the protein interaction network. Figure 10 shows the concrete scores of these hub genes. Finally, we chose the top 6 genes for use in further research.
VALIDATION OF DIAGNOSTIC VALUE OF HUB GENES:
To validate the diagnostic value of the top 6 hub genes obtained from the above analysis, we constructed ROC curves and calculated the corresponding area under the curve (AUC) of these gene expression levels in the NAFLD and AMI datasets. Figure 11 shows the results of NAFLD. The AUC for TLR2, LILRB2, CXCL1, FPR1, TLR4, and TYROBP in NAFLD patients and normal controls were 0.818 [95% confidence interval (CI), 0.619–1.000; P<0.05], 0.798 (95% CI, 0.584–1.000; P<0.05), 0.805 (95% CI, 0.558–1.000; P<0.05), 0.818 (95% CI, 0.610–1.000; P<0.05), 0.792 (95% CI, 0.563–1.000; P<0.05), and 0.831 (95% CI, 0.631–1.000; P<0.05). Figure 12 shows the ROC curve in AMI patients and non-AMI controls. The AUC for 6 hub genes in AMI were 0.762 [95% CI, 0.672–0.851; P<0.05], 0.798 (95% CI, 0.715–0.881; P<0.05), 0.742 (95% CI, 0.651–0.832; P<0.05), 0.748 (95% CI, 0.656–0.839; P<0.05), 0.758 (95% CI,0.666–0.849; P<0.05), and 0.677 (95% CI, 0.577–0.777; P<0.05) (Figure 12B).
Discussion
NAFLD and AMI are highly prevalent diseases around the world. A previous study showed that NAFLD patients are at increased risk of both hepatic complications and cardiac metabolic complications [22]. With longer course of disease, the risk increases gradually, so it is necessary to explore the molecular mechanisms in these 2 diseases and discover the early targets to prevent disease development [23].
In this study, through searching the datasets of NAFLD and AMI from GEO, we found 78 common DEGs between these diseases. We also performed GO enrichment and KEGG pathway enrichment analyses, and a PPI network was constructed to identify the top 10 hub genes from among the common DEGs. We chose 6 hub genes to verify diagnostic value in NAFLD and AMI patients (P<0.05). These genes may have an important ability to predict risk of AMI and T2DM.
Toll-like receptors (TLRs) belong to the family of pattern recognition receptors and play a vital role in recognizing bacterial and viral components, like lipopolysaccharides, bacterial DNA, and peptidoglycan [24,25]. Notably, among the TLRs family, toll-like receptor 2 (TLR2) and TLR4 have been reported to have a close relationship with pathogenesis of NAFLD [26]. A previous study indicated that the innate immune system plays an important role in the progression of NAFLD, and in this process, TRL4 is activated and participates in the disease sequence of steatosis, steatohepatitis, liver fibrosis, cirrhosis, and liver cancer [27]. In addition, Chiu et al. showed that, in a model of C57BL/6J mice fed with fresh fecal mixture obtained from NAFLD donors, the mice had higher TLR2 and TLR4 mRNA levels compared to control animals, which indicated these 2 factors participate in the gene regulation of disease [28]. A similar phenomenon can be seen in AMI pathogenesis, in which it was reported that TLR2/1 and TLR4 agonists mediate substantial direct platelet activation in AMI patients [29,30]. Furthermore, another study showed that TLR2 or TLR4 gene defects in mice can prevent cardiac remodeling, resulting in unchanged cardiac function and geometry after AMI [31].
Leukocyte immunoglobulin-like receptors subfamily B (LILRB) members are inhibitory receptors that can negatively regulate immune cell activation spontaneous by signaling intracellular immunoreceptor tyrosine-based inhibitory motifs (ITIMs) [32]. The LILRB2 gene locates in the region of human chromosome 19q13.4, and some immune-related receptors and genes have been reported [33]. Yan et al. studied expression of B cell-associated genes in people with AMI, stable angina, and healthy controls (n=20 per group), finding that LILRB2 was significantly overexpressed in the AMI group compared with the other 2 groups [34].
CXCL1 is a neutrophil chemokine that, when overexpressed in rat liver, results in hepatic dysfunction with neutrophil infiltration [35,36]. In a recent study, the expression of CXCL1 was significantly increased in the livers of NASH mice compared with mice that had simple hepatic steatosis [37]. Saeed et al. conducted an experiment on wild-type (WT) mice fed high-fat diets to form a NAFLD model, and showed that the gene expression of CXCL1 was higher in liver tissue than in normal controls [38]. Similarly, it has been reported that the level of CXCL1 is evaluated in AMI patients [39,40]. Pordel et al. also reported that the plasma levels of CXCL1 chemokine in AMI patients were notably higher than in healthy participants, possibly due to AMI activating an intense inflammatory response, and release of related pro-inflammatory cytokines such as CXCL1 and CXCL5 [41,42].
Formyl peptide receptor 1(FPR1) was first found in phagocytic leukocytes; it is a G protein-coupled 7 transmembrane cell surface receptor (GPCR). FPR1 has a variety of pathophysiologic functions such as inflammation, wound healing, glioblastoma progression, and defense against HCV infection [43,44]. The study of the relationship between NAFLD and FPR1 is incomplete, and previous research reported that liver cancer occurring at the end of NAFLD involves chronic inflammation, hepatocyte injury, and neovascularization [45]. Consist with other studies, Honda et al. induced liver necrosis in mice, finding that neutrophils quickly accumulated, but when FPR1 was inhibited, the neutrophil migration into the liver necrotic area was notably decreased [46]. Moreover, Zhou et al. studied the effect of FPR1 on ischemia/reperfusion injury in rats, showing that low expression of FPR1 depressed inflammation level, cardiomyocyte apoptosis, and ventricular remodeling [47]. Recent research also indicated that FPR1 thus might become a potential novel target in AMI treatment for improving the prognosis of MI patients [48,49].
Tyrosine kinase binding protein (TYROBP) is a regulatory protein of a variety of activated receptors in NK cells, which can bind to activated receptors in the form of noncovalent bonds, activate signal transduction, activate NK cells, and perform effective functions [50]. In a TYROBP gene knockout model, the level of pro-inflammatory cytokines was decreased, such as TNF-α, IL-10, IL-6, and MCP-1, and TYROBP was found to synthesize lipopolysaccharide and induce the production of pro-inflammatory cytokines mediated by toll-like receptor [51]. NK cells participate in the pathophysiological process of NAFLD and AMI [52,53]; suggesting that TYROBP may also play an important role through NK cells in these diseases, but further research on this is needed.
We also assessed associated gene biological functions and pathways, such as inflammatory reactions of GO terms and immune system diseases pathways, IL-17 signaling pathway, and NF-kappa B signaling pathway, which were reported to be closely involved in both diseases [54–56].
Conclusions
In this study, 2 AMI datasets and 1 NAFLD dataset were downloaded from GEO. After data selection, 78 intersection genes were obtained from these datasets when patient samples were compared to normal ones. Through GO and KEGG enrichment analyses, inflammation-related biological functions and pathways were obtained. Using PPI network construction, the hub genes TLR2, LILRB2, CXCL1, FPR1, TLR4, and TYROBP were selected and their diagnostic values were validated by SPSS data analysis. The outcomes of our study may provide potential targets for the prevention and treatment of NAFLD and AMI.
However, there are some limitations of our study. First, the DEGs screened by bioinformatics method can predict the occurrence of NAFLD and AMI, and our research team is working on
Figures
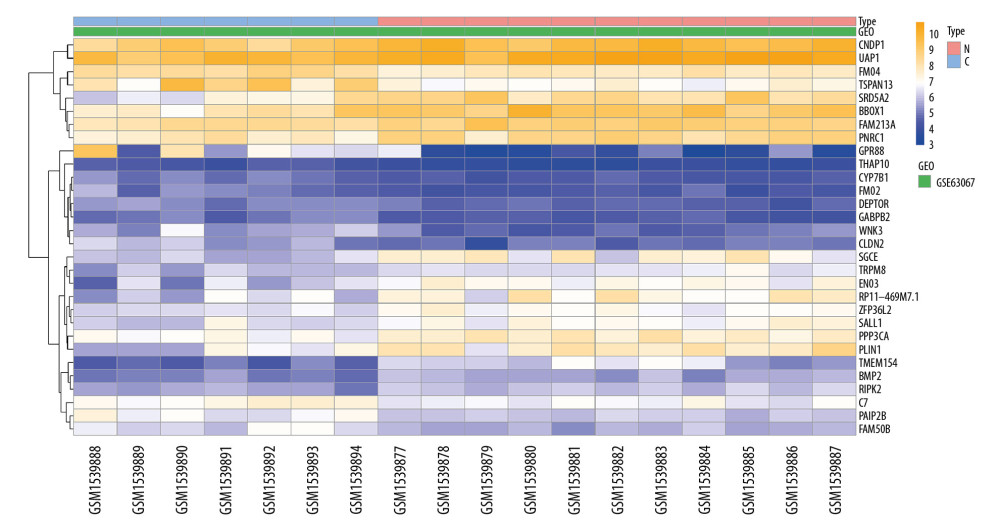 Figure 1. Heatmap showing the expression changes in non-alcoholic fatty liver disease (NAFLD). N – NAFLD; C – control.
Figure 1. Heatmap showing the expression changes in non-alcoholic fatty liver disease (NAFLD). N – NAFLD; C – control. 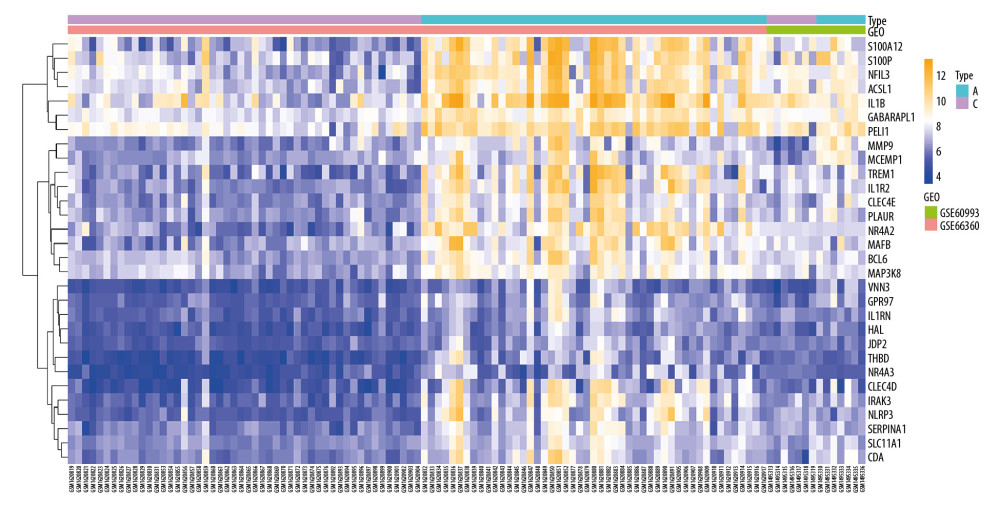 Figure 2. Heatmap showing the expression changes in acute myocardial infarction (AMI). A – AMI; C – control.
Figure 2. Heatmap showing the expression changes in acute myocardial infarction (AMI). A – AMI; C – control. 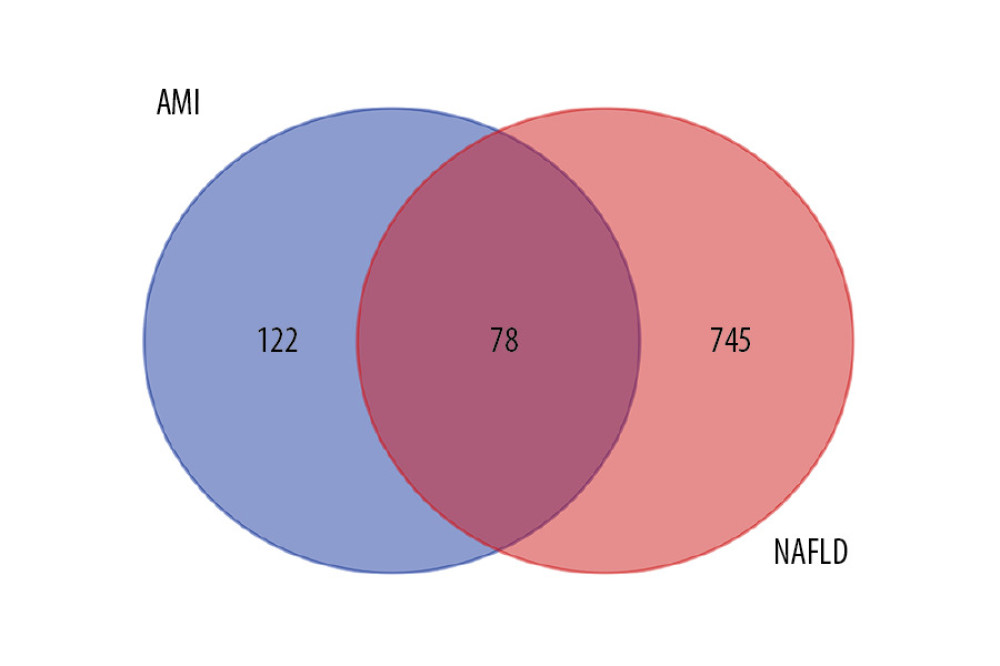 Figure 3. Venn diagram of intersecting common genes identified by differential genes (DEGs) from NAFLD and AMI.
Figure 3. Venn diagram of intersecting common genes identified by differential genes (DEGs) from NAFLD and AMI. 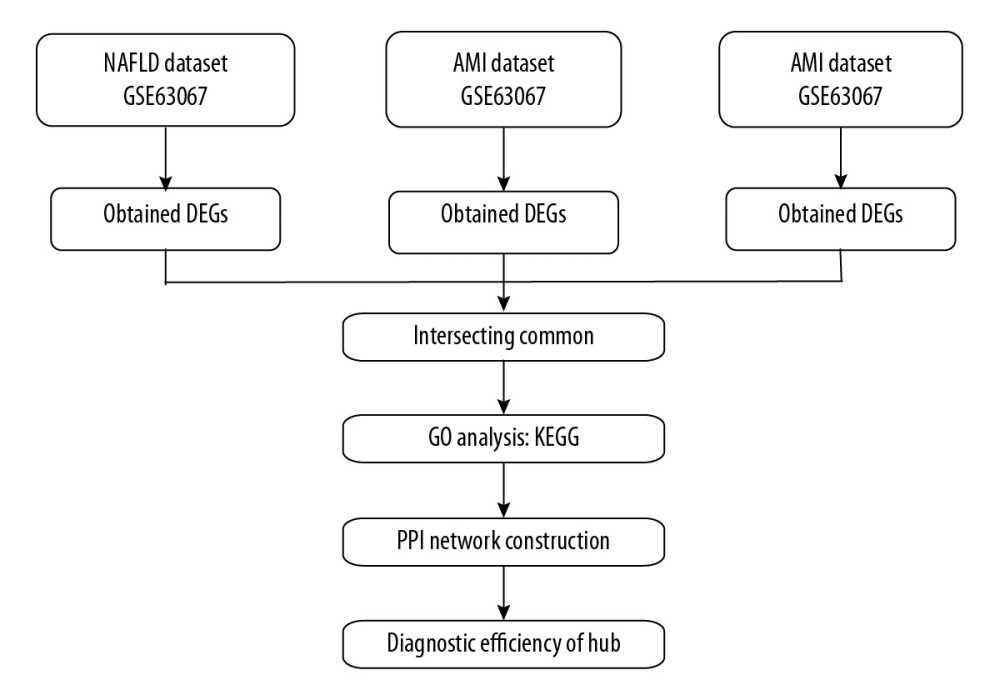 Figure 4. Flow diagram of study.
Figure 4. Flow diagram of study. 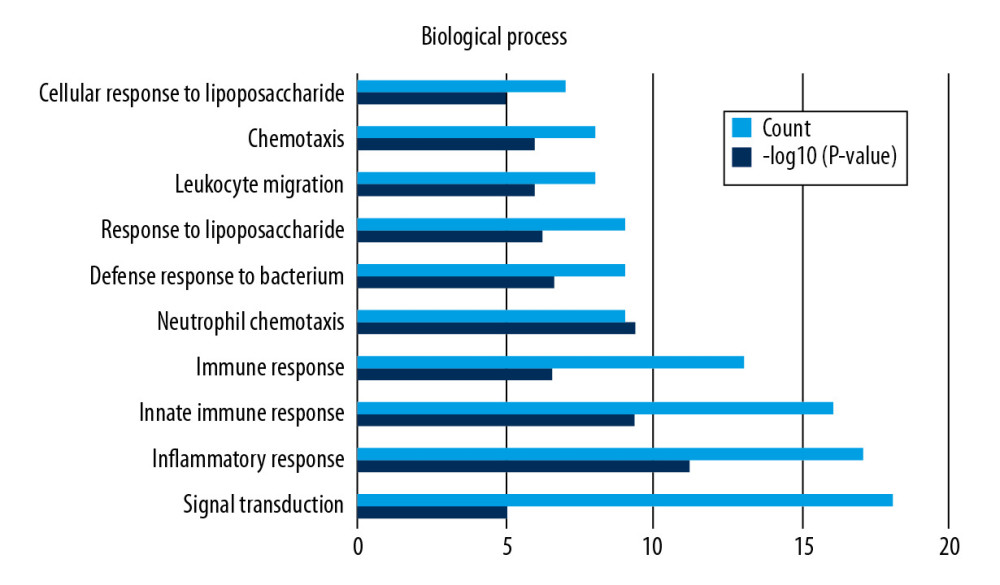 Figure 5. GO analysis of biological processes (BP).
Figure 5. GO analysis of biological processes (BP). 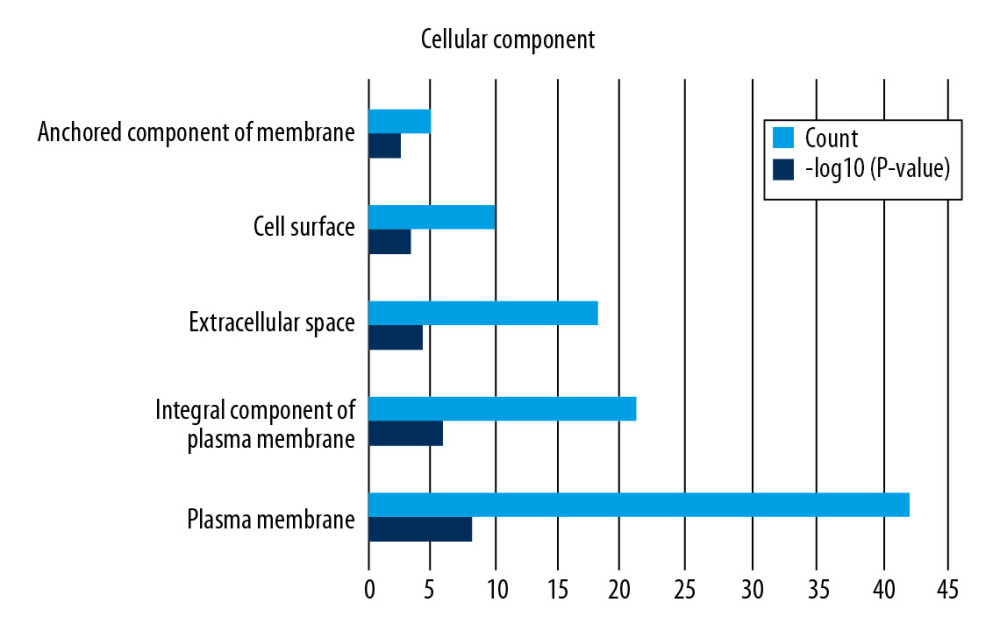 Figure 6. GO analysis of cellular component (CC).
Figure 6. GO analysis of cellular component (CC). 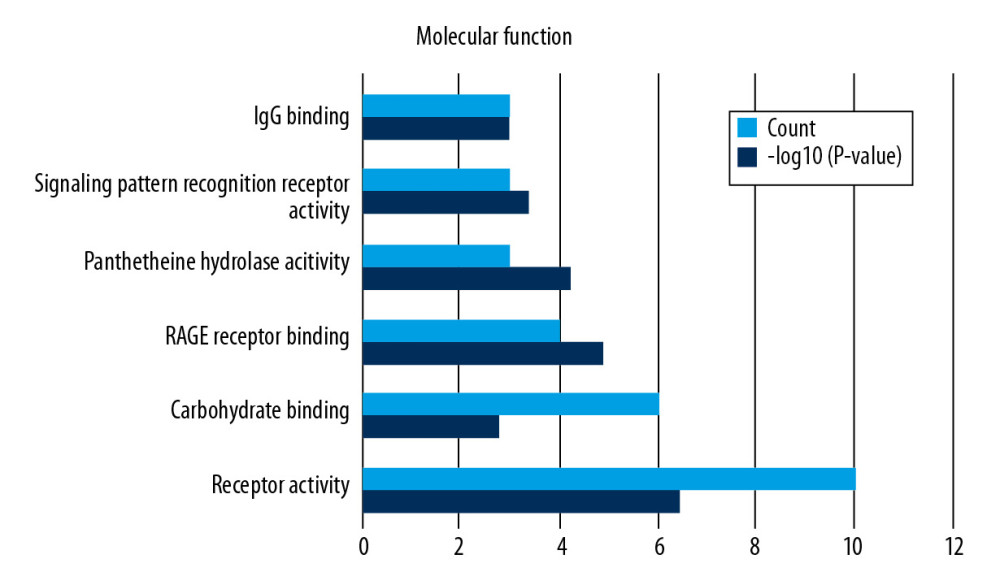 Figure 7. GO analysis of molecular function (MF).
Figure 7. GO analysis of molecular function (MF). 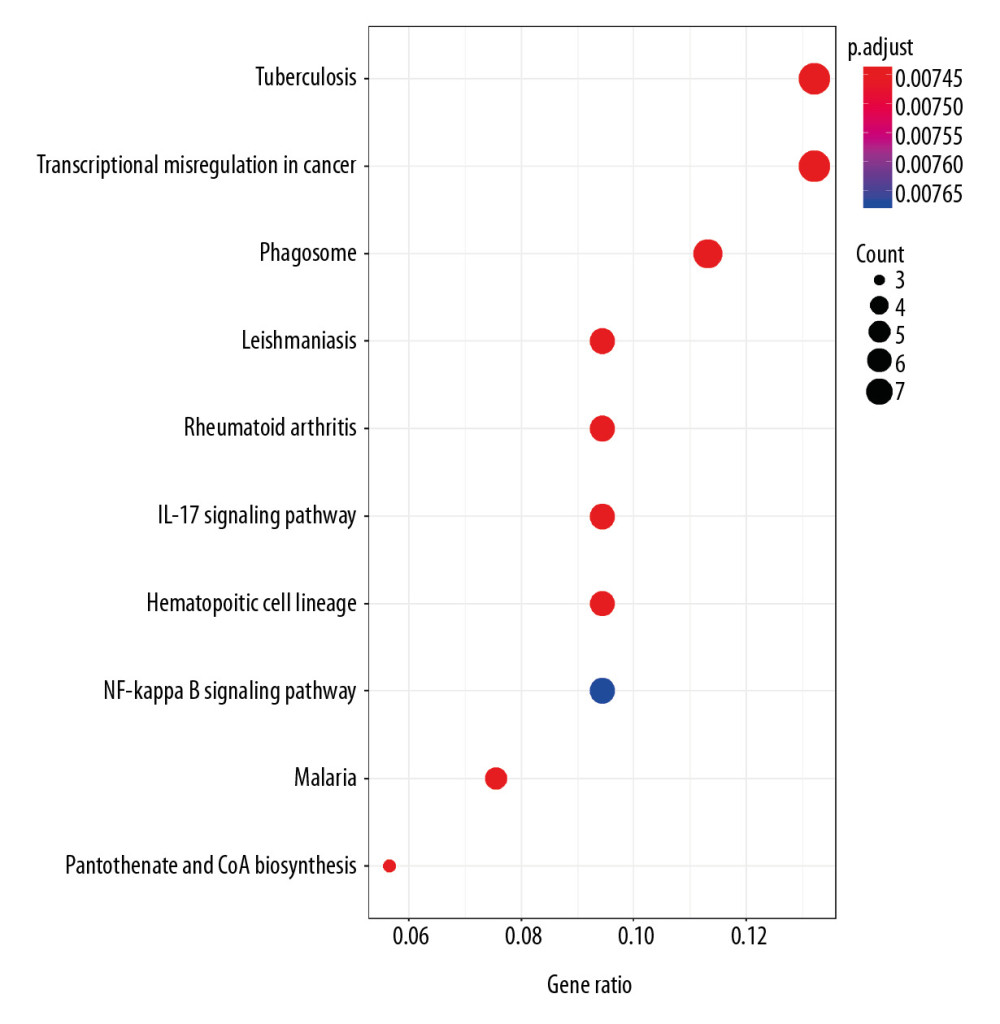 Figure 8. KEGG pathway enrichment analysis of common genes.
Figure 8. KEGG pathway enrichment analysis of common genes. 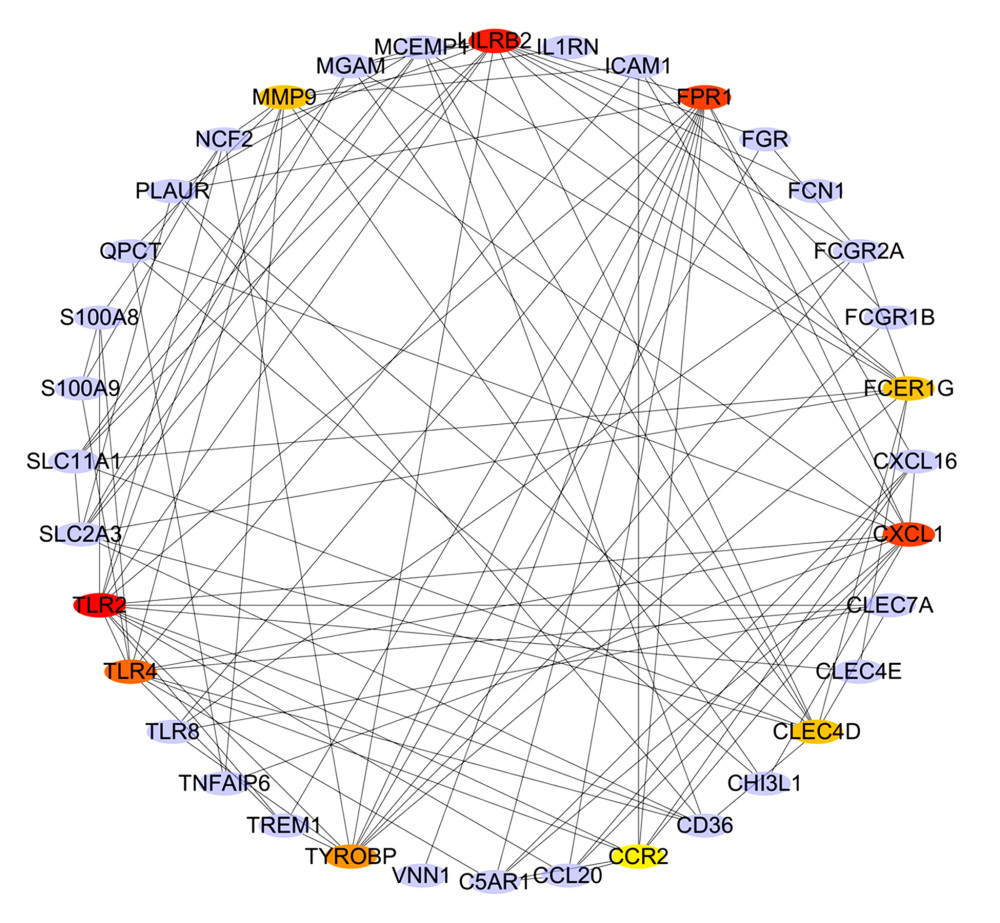 Figure 9. PPI network construction and the hub genes in different color.
Figure 9. PPI network construction and the hub genes in different color. 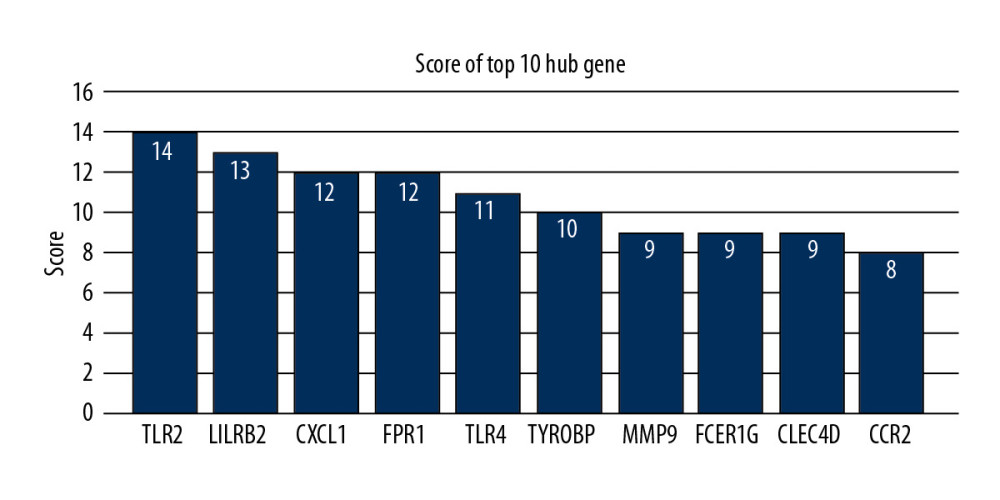 Figure 10. The accordance percentage of hub genes.
Figure 10. The accordance percentage of hub genes. 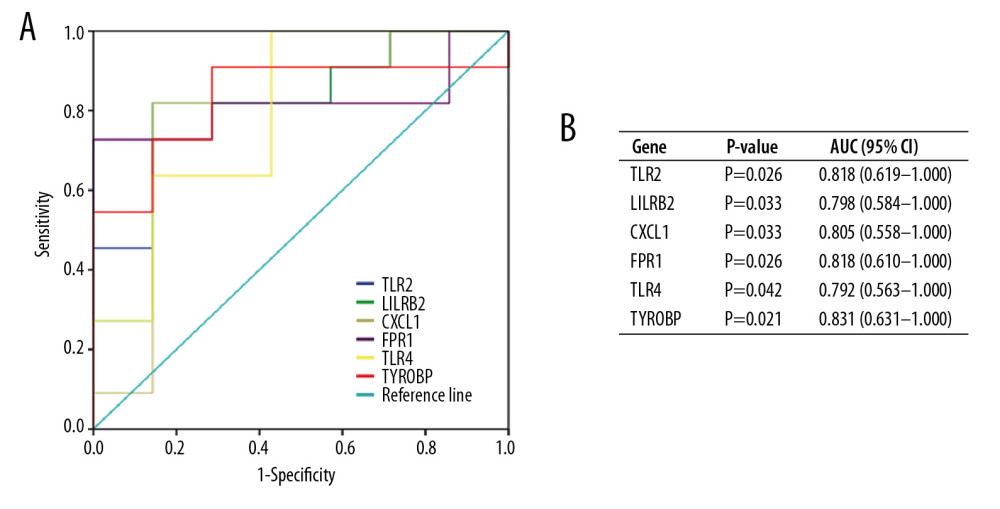 Figure 11. Diagnostic value of top 6 hub genes with ROC curves in NAFLD. (A) Analysis with ROC curves. (B) Specific value of diagnosis efficiency.
Figure 11. Diagnostic value of top 6 hub genes with ROC curves in NAFLD. (A) Analysis with ROC curves. (B) Specific value of diagnosis efficiency. 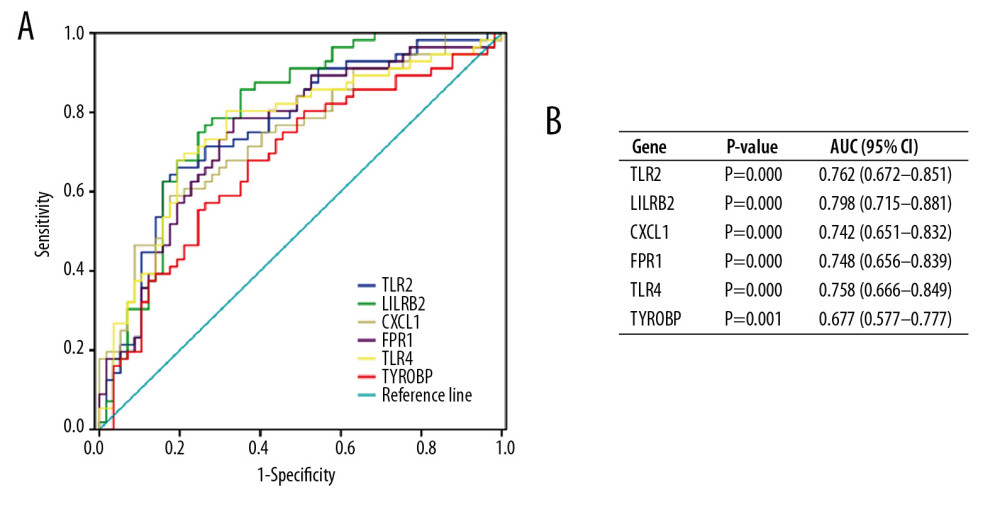 Figure 12. Diagnostic value of top 6 hub genes with ROC curves in AMI. (A) Analysis with ROC curves. (B) Specific value of diagnosis efficiency.
Figure 12. Diagnostic value of top 6 hub genes with ROC curves in AMI. (A) Analysis with ROC curves. (B) Specific value of diagnosis efficiency. References
1. Marra F, Svegliati-Baroni G, Lipotoxicity and the gut-liver axis in NASH pathogenesis: J Hepatol, 2018; 68; 280-95
2. Younossi ZM, Koenig AB, Abdelatif D, Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes: Hepatology, 2016; 64; 73-84
3. Buzzetti E, Pinzani M, Tsochatzis EA, The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD): Metab Clin Exp, 2016; 65; 1038-48
4. Estes C, Razavi H, Loomba R, Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease: Hepatology, 2018; 67; 123-33
5. Younossi ZM, Tampi R, Priyadarshini M, Burden of illness and economic model for patients with nonalcoholic steatohepatitis in the United States: Hepatology, 2019; 69; 564-72
6. Mozaffarian D, Benjamin EJ, GO ASWriting Group Members, Heart disease and stroke statistics – 2016 update: A Report from the American Heart Association: Circulation, 2016; 133; e38-360
7. Wołkow PP, Drabik L, Totoń-Żurańska J, Polymorphism in the chemokine receptor 7 gene (CCR7) is associated with previous myocardial infarction in patients undergoing elective coronary angiography: Int J Immunogenet, 2016; 43; 218-25
8. Kazuhiro N, Tetsuya M, Yajing M, A new therapeutic modality for acute myocardial infarction: nanoparticle-mediated delivery of pitavastatin induces cardioprotection from ischemia-reperfusion injury via activation of PI3K/Akt pathway and anti-inflammation in a rat model: PLoS One, 2015; 10; e0132451
9. European Association for the Study of the Liver (EASL), European Association for the Study of Diabetes (EASD), European Association for the Study of Obesity (EASO), EASL-EASD-EASO Clinical Practice Guidelines for the management of non-alcoholic fatty liver disease: J Hepatol, 2016; 64; 1388-402
10. Donghee K, Ray KW, Kim HJ, Thernau TM, Association between noninvasive fibrosis markers and mortality among adults with nonalcoholic fatty liver disease in the United States: Hepatology, 2013; 57; 1357-65
11. Sinn DH, Kang K, Chang Y, Non-alcoholic fatty liver disease and the incidence of myocardial infarction: A cohort study: J Gastroenterol Hepatol; 2019 [Epub ahead of print]
12. Targher G, Byrne CD, Lonardo A, Non-alcoholic fatty liver disease and risk of incident cardiovascular disease: A meta-analysis: J Hepatol, 2016; 65; 589-600
13. Alexander M, Loomis AK, van der Lei J, Non-alcoholic fatty liver disease and risk of incident acute myocardial infarction and stroke: Findings from matched cohort study of 18 million European adults: BMJ, 2019; 367; l5367
14. Liu K, Kang M, Li J, Prognostic value of the mRNA expression of members of the HSP90 family in non-small cell lung cancer: Exp Ther Med, 2019; 17; 2657-65
15. Ritchie ME, Phipson B, Wu D, limma powers differential expression analyses for RNA-sequencing and microarray studies: Nucleic Acids Res, 2015; 43; e47
16. Gene Ontology Consortium, The Gene Ontology (GO) project in 2006: Nucleic Acids Res, 2006; 34; D322-26
17. Kanehisa M, Sato Y, Kawashima M, KEGG as a reference resource for gene and protein annotation: Nucleic Acids Res, 2016; 44; D457-62
18. Szklarczyk D, Franceschini A, Wyder S, STRING v10: Protein-protein interaction networks, integrated over the tree of life: Nucleic Acids Res, 2015; 43; D447-52
19. Su G, Morris JH, Demchak B, Biological network exploration with Cytoscape 3: Curr Protoc Bioinformatics, 2014; 47; 8.13.1-24
20. Yang Y, Lin J, Ma Z, Potential roles of microRNAs and their target genes in human multiple myeloma: Eur J Haematol, 2017; 99; 178-85
21. Chin CH, Chen SH, Wu HH, cytoHubba: Identifying hub objects and sub-networks from complex interactome: BMC Syst Biol, 2014; 8(Suppl 1); S11
22. Bhatia LS, Curzen NP, Calder PC, Non-alcoholic fatty liver disease: A new and important cardiovascular risk factor?: Eur Heart J, 2012; 33; 1190-200
23. Leung JC, Loong TC, Wei L, Histological severity and clinical outcomes of nonalcoholic fatty liver disease in nonobese patients: Hepatology, 2017; 65; 54-64
24. Aderem A, Ulevitch RJ, Toll-like receptors in the induction of the innate immune response: Nature, 2000; 406; 782-87
25. Beutler BA, TLRs and innate immunity: Blood, 2009; 113; 1399-407
26. Kapil S, Duseja A, Sharma BK, CD14Genetic polymorphism in gene, a co-receptor of TLR4 associated with non-alcoholic fatty liver disease: World J Gastroenterol, 2016; 22; 9346-55
27. Lopes JA, Borges-Canha M, Pimentel-Nunes P, Innate immunity and hepatocarcinoma: Can toll-like receptors open the door to oncogenesis?: World J Hepatol, 2016; 8; 162-82
28. Chiu CC, Ching YH, Li YP, Nonalcoholic fatty liver disease is exacerbated in high-fat diet-fed gnotobiotic mice by colonization with the gut microbiota from patients with nonalcoholic steatohepatitis: Nutrients, 2017; 9(11) pii: E1220
29. Hally KE, La Flamme AC, Larsen PD, Platelet Toll-like receptor (TLR) expression and TLR-mediated platelet activation in acute myocardial infarction: Thromb Res, 2017; 158; 8-15
30. Rivadeneyra L, Carestia A, Etulain J, Regulation of platelet responses triggered by Toll-like receptor 2 and 4 ligands is another non-genomic role of nuclear factor-kappaB: Thromb Res, 2014; 133; 235-43
31. Shishido T, Nozaki N, Takahashi H, Central role of endogenous Toll-like receptor-2 activation in regulating inflammation, reactive oxygen species production, and subsequent neointimal formation after vascular injury: Biochem Biophys Res Commun, 2006; 345; 1446-53
32. Brown D, Trowsdale J, Allen R, The LILR family: Modulators of innate and adaptive immune pathways in health and disease: Tissue Antigens, 2004; 64; 215-25
33. Hirayasu K, Arase H, Functional and genetic diversity of leukocyte immunoglobulin-like receptor and implication for disease associations: J Hum Genet, 2015; 60; 703-8
34. Yan W, Song H, Jiang J, Characteristics of B cell-associated gene expression in patients with coronary artery disease: Mol Med Rep, 2016; 13; 4113-21
35. Moser B, Clark-Lewis I, Zwahlen R, Neutrophil-activating properties of the melanoma growth-stimulatory activity: J Exp Med, 1990; 171; 1797-802
36. Maher JJ, Scott MK, Saito JM, Adenovirus-mediated expression of cytokine-induced neutrophil chemoattractant in rat liver induces a neutrophilic hepatitis: Hepatology, 1997; 25; 624-30
37. Semba T, Nishimura M, Nishimura S, The FLS (fatty liver Shionogi) mouse reveals local expressions of lipocalin-2, CXCL1 and CXCL9 in the liver with non-alcoholic steatohepatitis: BMC Gastroenterol, 2013; 13; 120
38. Saeed WK, Jun DW, Jang K, Mismatched effects of receptor interacting protein kinase-3 on hepatic steatosis and inflammation in non-alcoholic fatty liver disease: World J Gastroenterol, 2018; 24; 5477-90
39. Barry SP, Ounzain S, McCormick J, Enhanced IL-17 signalling following myocardial ischaemia/reperfusion injury: Int J Cardiol, 2013; 163; 326-34
40. Ivey CL, Williams FM, Collins PD, Neutrophil chemoattractants generated in two phases during reperfusion of ischemic myocardium in the rabbit. Evidence for a role for C5a and interleukin-8: J Clin Invest, 1995; 95; 2720-28
41. Pordel S, Sajedi Khanian M, Karimi MH, Plasma CXCL1 levels and TRAF3IP2 variants in patients with myocardial infarction: J Clin Lab Anal, 2018; 32(6); e22402
42. Frangogiannis NG, The inflammatory response in myocardial injury, repair, and remodelling: Nat Rev Cardiol, 2014; 11; 255-65
43. Liu M, Chen K, Yoshimura T, Formylpeptide receptors mediate rapid neutrophil mobilization to accelerate wound healing: PLoS One, 2014; 9; e90613
44. Lin Q, Fang D, Hou X, HCV peptide (C5A), an amphipathic α-helical peptide of hepatitis virus C, is an activator of N-formyl peptide receptor in human phagocytes: J Immunol, 2011; 186; 2087-94
45. Bishayee A, The role of inflammation and liver cancer: Adv Exp Med Biol, 2014; 816; 401-35
46. Honda M, Takeichi T, Hashimoto S, Intravital imaging of neutrophil recruitment reveals the efficacy of FPR1 blockade in hepatic ischemia-reperfusion injury: J Immunol, 2017; 198; 1718-28
47. Zhou QL, Teng F, Zhang YS, FPR1 gene silencing suppresses cardiomyocyte apoptosis and ventricular remodeling in rats with ischemia/reperfusion injury through the inhibition of MAPK signaling pathway: Exp Cell Res, 2018; 370; 506-18
48. Perretti M, Leroy X, Bland EJ, Resolution pharmacology: Opportunities for therapeutic innovation in inflammation: Trends Pharmacol Sci, 2015; 36; 737-55
49. Qin CX, May LT, Li R, Small-molecule-biased formyl peptide receptor agonist compound 17b protects against myocardial ischaemia-reperfusion injury in mice: Nat Commun, 2017; 8; 14232
50. Tomasello E, Vivier E, KARAP/DAP12/TYROBP: Three names and a multiplicity of biological functions: Eur J Immunol, 2005; 35; 1670-77
51. Turnbull IR, McDunn JE, Takai T, DAP12 (KARAP) amplifies inflammation and increases mortality from endotoxemia and septic peritonitis: J Exp Med, 2005; 202; 363-69
52. Cuff AO, Sillito F, Dertschnig S, The obese liver environment mediates conversion of NK cells to a less cytotoxic ILC1-like phenotype: Front Immunol, 2019; 10; 2180
53. Rakic M, Persic V, Kehler T, Possible role of circulating endothelial cells in patients after acute myocardial infarction: Med Hypotheses, 2018; 117; 42-46
54. Lu Y, Lin Y, Huang X, Oxaliplatin aggravates hepatic oxidative stress, inflammation and fibrosis in a non-alcoholic fatty liver disease mouse model: Int J Mol Med, 2019; 43; 2398-408
55. Vonghia L, Magrone T, Verrijken A, Peripheral and hepatic vein cytokine levels in correlation with non-alcoholic fatty liver disease (NAFLD)-related metabolic, histological, and haemodynamic features: PLoS One, 2015; 10; e0143380
56. Chang H-Y, Li X, Tian Y, Telmisartan reduces arrhythmias through increasing cardiac connexin43 by inhibiting IL-17 after myocardial infarction in rats: Eur Rev Med Pharmacol Sci, 2017; 21; 5283-89
Figures
 Figure 1. Heatmap showing the expression changes in non-alcoholic fatty liver disease (NAFLD). N – NAFLD; C – control.
Figure 1. Heatmap showing the expression changes in non-alcoholic fatty liver disease (NAFLD). N – NAFLD; C – control. Figure 2. Heatmap showing the expression changes in acute myocardial infarction (AMI). A – AMI; C – control.
Figure 2. Heatmap showing the expression changes in acute myocardial infarction (AMI). A – AMI; C – control. Figure 3. Venn diagram of intersecting common genes identified by differential genes (DEGs) from NAFLD and AMI.
Figure 3. Venn diagram of intersecting common genes identified by differential genes (DEGs) from NAFLD and AMI. Figure 4. Flow diagram of study.
Figure 4. Flow diagram of study. Figure 5. GO analysis of biological processes (BP).
Figure 5. GO analysis of biological processes (BP). Figure 6. GO analysis of cellular component (CC).
Figure 6. GO analysis of cellular component (CC). Figure 7. GO analysis of molecular function (MF).
Figure 7. GO analysis of molecular function (MF). Figure 8. KEGG pathway enrichment analysis of common genes.
Figure 8. KEGG pathway enrichment analysis of common genes. Figure 9. PPI network construction and the hub genes in different color.
Figure 9. PPI network construction and the hub genes in different color. Figure 10. The accordance percentage of hub genes.
Figure 10. The accordance percentage of hub genes. Figure 11. Diagnostic value of top 6 hub genes with ROC curves in NAFLD. (A) Analysis with ROC curves. (B) Specific value of diagnosis efficiency.
Figure 11. Diagnostic value of top 6 hub genes with ROC curves in NAFLD. (A) Analysis with ROC curves. (B) Specific value of diagnosis efficiency. Figure 12. Diagnostic value of top 6 hub genes with ROC curves in AMI. (A) Analysis with ROC curves. (B) Specific value of diagnosis efficiency.
Figure 12. Diagnostic value of top 6 hub genes with ROC curves in AMI. (A) Analysis with ROC curves. (B) Specific value of diagnosis efficiency. In Press
07 Mar 2024 : Clinical Research
Knowledge of and Attitudes Toward Clinical Trials: A Questionnaire-Based Study of 179 Male Third- and Fourt...Med Sci Monit In Press; DOI: 10.12659/MSM.943468
08 Mar 2024 : Animal Research
Modification of Experimental Model of Necrotizing Enterocolitis (NEC) in Rat Pups by Single Exposure to Hyp...Med Sci Monit In Press; DOI: 10.12659/MSM.943443
18 Apr 2024 : Clinical Research
Comparative Analysis of Open and Closed Sphincterotomy for the Treatment of Chronic Anal Fissure: Safety an...Med Sci Monit In Press; DOI: 10.12659/MSM.944127
08 Mar 2024 : Laboratory Research
Evaluation of Retentive Strength of 50 Endodontically-Treated Single-Rooted Mandibular Second Premolars Res...Med Sci Monit In Press; DOI: 10.12659/MSM.944110
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952