11 July 2020: Database Analysis
Underlying Mechanism of Insulin Resistance: A Bioinformatics Analysis Based on Validated Related-Genes from Public Disease Databases
Peng Gao1ACE, Yan Hu1BE, Junyan Wang1BF, Yinghua Ni1C, Zhengyi Zhu1D, Huijuan Wang1C, Jufei Yang1D, Lingfei Huang1F, Luo Fang1AEG*DOI: 10.12659/MSM.924334
Med Sci Monit 2020; 26:e924334
Abstract
BACKGROUND: The underlying mechanism of insulin resistance is complex; bioinformatics analysis is used to explore the mechanism based differential expression genes (DEGs) obtained from omics analysis. However, the expression and role of most DEGs involved in bioinformatics analysis are invalidated. This study aimed to disclose the mechanism of insulin resistance via bioinformatics analysis based on validated insulin resistance-related genes (IRRGs) collected from public disease-gene databases.
MATERIAL AND METHODS: IRRGs were collected from 4 disease databases including NCBI-Gene, CTD, RGD, and Phenopedia. GO and KEGG analysis of IRRGs were performed by DAVID. Then, the STRING database was employed to construct a protein–protein interaction (PPI) network of IRRGs. The module analysis and hub genes identification were carried out by MCODE and cytoHubba plugin of Cytoscape based on the primary PPI network, respectively.
RESULTS: A total of 1195 IRRGs were identified. Response to drug, hypoxia, insulin, positive regulation of transcription from RNA polymerase II promoter, cell proliferation, inflammatory response, negative regulation of apoptotic process, glucose homeostasis, cellular response to insulin stimulus, and aging were proposed as the crucial functions related to insulin resistance. Ten insulin resistance-related pathways included the pathways of insulin resistance, pathways in cancer, adipocytokine, prostate cancer, PI3K-Akt, insulin, AMPK, HIF-1, prolactin, and pancreatic cancer signaling pathway were revealed. INS, AKT1, IL-6, TP53, TNF, VEGFA, MAPK3, EGFR, EGF, and SRC were identified as the top 10 hub genes.
CONCLUSIONS: The current study presented a landscape view of possible underlying mechanism of insulin resistance by bioinformatics analysis based on validated IRRGs.
Keywords: database, Genes, vif, Insulin Resistance, Computational Biology, Databases, Genetic, Gene Expression Profiling, gene ontology, Gene Regulatory Networks, Protein Interaction Maps, Software
Background
Insulin resistance is a pathological condition with impaired sensitivity to insulin in target tissues including liver, heart, muscle, and adipose tissue [1]. Insulin resistance is strongly related to most metabolic disorders, such as obesity, dyslipidemia, hypertension, atherosclerosis, and endothelial dysfunction [2–5], and it is one of the key contributors to diabetes, cardiovascular disease, metabolic syndrome, and Alzheimer’s disease [6–9]. The incidence of insulin resistance and related complications is still rising [10]. Thus, it is essential to explore the underlying mechanism of insulin resistance, which may be conducive to guide relevant clinical diagnosis and therapy.
The underlying mechanisms of insulin resistance include multi-fields crossed inflammation, endoplasmic reticulum stress, oxidative stress, and mitochondrial dysfunction [11–14]. Accordingly, hundreds of key molecules have been identified [15–17], such as insulin receptor substrates (IRSs), GLUT4, protein-tyrosine phosphatase 1B (PTP1B), tumor necrosis factor-α (TNF-α), interleukins (ILs), and so on. Signaling pathways like PI3K-Akt, ERK, and adipocytokine play crucial roles in the development of insulin resistance [18–20]. The underlying mechanism of insulin resistance is intricate, and it is hard to get a clear overview by looking at a few molecular or single-pathway studies.
Recently, bioinformatics analysis as an interdisciplinary field has been viewed as an efficient tool in the study of complex mechanisms [21]. Two studies of insulin resistance were reported based on differential expressed genes (DEGs) between insulin sensitive and insulin resistant tissues extracted from the Gene Expression Omnibus (GEO) [22,23]. One study disclosed insulin resistance related transcription factors, including ETS1, AR, ESR1, and Myc [22], and the other revealed functions, signal pathway, and hub genes significantly related to insulin resistance [23]. The DEGs obtained from omics analysis have been commonly used in an overwhelming majority of bioinformatics analysis. However, DEGs require further validation of expression level by quantitative real-time PCR (qPCR) [24] and identification of their real roles in insulin resistance: acted as regulators of insulin resistance, or only genes regulated by insulin resistance. Although validated DEGs based analysis could provide more solid evidence, there have been no validated DEGs based analysis reported, as it is difficult to validate massive DEGs. Recently, several databases have collected disease-related genes that provide an alternative insulin resistance-related gene (IRRG) collection. Therefore, we developed a bioinformatics analysis based on validated IRRGs extracted from published disease-gene association databases to disclose the underlying mechanism of insulin resistance. In the present study, we collected IRRGs from 4 disease databases including The National Center for Biotechnology Information’s Gene database (NCBI-Gene) [25], the Comparative Toxicogenomics Database (CTD) [26], the Rat Genome Database (RGD) [27] and Phenopedia [28]. Our study aimed to provide comprehensive understanding of biological functions, pathways, and key molecules of insulin resistance by bioinformatics analysis.
Material and Methods
IRRGS COLLECTION:
IRRGs were collected from the databases of NCBI-Gene [29], CTD [30], RGD [31], and Phenopedia [32] on January 21, 2019. The IRRGs were searched from NCBI-Gene database with the following text words [(insulin resistance OR insulin resistant) AND Homo sapiens], from RGD with a disease category of insulin resistance, from CTD database of the genes with a curated association (marker/mechanism) to insulin resistance, and from Phenopedia of the genes linked to insulin resistance disease by at least 2 publications. All the IRRGs from the 4 databases were aggregated and duplicated genes were removed.
FUNCTIONAL ANNOTATION AND SIGNALING PATHWAY ANALYSIS:
Gene ontology (GO) analysis of cellular component (CC), molecular function (MF), and biological process (BP), and the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of signaling pathway enrichment were performed via the Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resources 6.8 [33,34] based on IRRGs. P<0.05 was set as statistically significant.
PROTEIN–PROTEIN INTERACTION (PPI) NETWORK CONSTRUCTION, MODULE ANALYSIS AND HUB GENES:
The IRRGs were mapped in the Search Tool for the Retrieval of Interacting Genes database (STRING) [35,36], and the interactive relationships with confidence score ≥0.4 were identified. Both primary network contained overall protein–protein interactions (PPIs) and sub-clusters were achieved by using Cytoscape software package (version 3.6.1, U.S. National Institute of General Medical Sciences) [37] plus Molecular Complex Detection (MCODE) plugin at degree cutoff of 2, node score cutoff of 0.2, the k-core value of 2, and the maximum depth of 100. Moreover, DAVID were employed to GO analyses in the modules. The hub genes were identified by degree method [38] using the cytoHubba plugin of the Cytoscape software.
Results
COLLECTION OF IRRGS:
A total of 1195 IRRGs were obtained from the 4 databases. The flow chart of collecting IRRGs is shown in Figure 1.
GO:
The biological process of response to drug, hypoxia, insulin, positive regulation of transcription from RNA polymerase II promoter, cell proliferation, inflammatory response, negative regulation of apoptotic process, glucose homeostasis, cellular response to insulin stimulus, and aging were main terms of BP. The genes were primarily enriched in cellular components of extracellular space, extracellular region, cytosol, cell surface, and membrane raft. Protein, enzyme, receptor, transcription factor binding, and protein homodimerization activity were among the top enriched in MF. The top 20 enriched terms of GO analysis are shown in Figure 2A–2C.
SIGNALING PATHWAYS:
There were 138 signaling pathways enriched IRRGs from KEGG. The main pathways involved in insulin resistance, pathways in cancer, adipocytokine signaling pathway, prostate cancer, PI3K-Akt, insulin, AMPK, HIF-1, prolactin signaling pathway, and pancreatic cancer. The top 20 terms of KEGG pathway are shown in Figure 2D.
PPI NETWORKS:
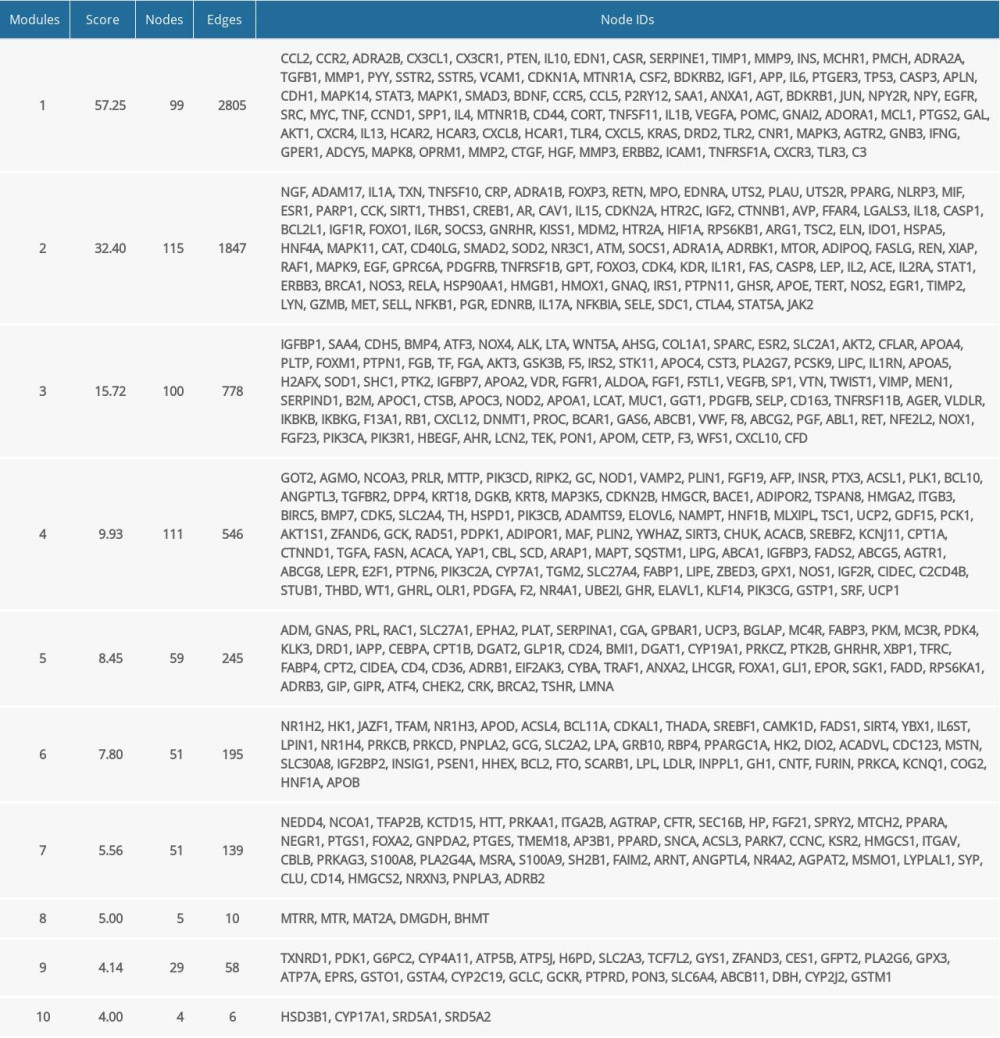
The PPI network involved 1119 nodes and 15 308 edges. Then, MCODE was performed to detect significant modules in this PPI network, and there were listed the top 10 modules of the networks in Table 1. BP of the top 4 modules are displayed. The results are shown in Figure 3. The most significant BP enriched in module 1 was inflammatory response. Other significant BP included positive regulation of nitric oxide biosynthetic process, and ERK1 and ERK2 cascade. In module 2, negative regulation of apoptotic process, response to hypoxia, and signal transduction were documented. The BP in module 3 were linked to metabolism, included high-density lipoprotein particle remodeling, reverse cholesterol transport, and lipoprotein metabolic process. Module 4 was linked to negative regulation of apoptotic process, response to nutrient, and cellular response to insulin stimulus.
HUB GENES:
Insulin (INS), AKT serine/threonine kinase 1 (AKT1), interleukin 6 (IL-6), tumor protein P53 (TP53), TNF, vascular endothelial growth factor A (VEGFA), mitogen-activated protein kinase 3 (MAPK3), epidermal growth factor receptor (EGFR), epidermal growth factor (EGF), and SRC proto-oncogene, non-receptor tyrosine kinase (SRC) were suggested as the top 10 hub genes (Figure 4).
Discussion
In this study, we explored the key molecules, functions, and pathways to display a comprehensive molecular mechanism of insulin resistance based on IRRGs collected from public disease databases. Although basic researches of insulin resistance mechanism were prevalent, the responding bioinformatics analysis reported by Yang et al. in 2014 [22] and Zhang et al. in 2019 [23]. In the previous studies, bioinformatics analysis was performed to construct regulatory networks and identify molecular biomarkers for insulin resistance based on DEGs shared in the GEO databases. Several transcription factors or hub genes were identified as potential biomarkers of insulin resistance. The 2 papers provided excellent examples for understanding of insulin resistance and exploring the biomarkers. For the limitation of the previous studies that most DEGs involved in the analysis were without validation of either differential expression or correlation to insulin resistance, we conducted the bioinformatics analysis based on validated IRRGs from public diseases database in the present study.
There were dramatically boosted research studies focused on IRRGs that provide a huge resource for bioinformatics analysis. However, how to comprehensively collect IRRGs from overload information has been challenging work. For our study, the 4 databases, including NCBI-Gene [25], CTD [26], RGD [27], and Phenopedia [28] were integrated. The 4 databases manually or automatically retrieve disease related genes from published literature. Moreover, these databases additionally included the genes related to human diseases from other databases, such as GeneCards and OMIM, to make sure comprehensive coverage of genes. A total of 1195 IRRGs were involved in the present study. The size of genes was moderate and comparable to the reported study by Yang et al. [22] but larger than Zhang et al. [23]. On this basis, we sketched the landscape of biologic feature and got the key molecules of IRRGs.
The biological function related to insulin stimulus, response and secretion, hypoxia, apoptotic process, cell proliferation, inflammatory response, and response to lipopolysaccharide and glucocorticoid, were enriched as the crucial functions of insulin resistance. Recently, inflammatory response, hypoxia, and apoptosis were proposed as hot spots for publications. Hypoxia is one of the key factors that induces inflammatory and apoptosis to enhance insulin resistance [39,40]. During the progressive development of obesity, the adipocytes became hypoxic and hypoxia-inducible factor 1 (HIF-1), nuclear factor kappaB (NF-κB) and adipocytokine, such as TNF-α, IL-6, and transforming growth factor-beta (TGF-β), are upregulated [41,42]. Also, the induction of adipocyte apoptosis is an alternative mechanism to explain hypoxia which contributes to inflammation in obesity [43]. Besides, hypoxia inhibits glucose uptake by reducing insulin receptor-β (IR-β) and IRS-1 to enhance insulin resistance [43]. Cell apoptosis, such as p53 induced apoptosis of adipocyte and fatty acid induced apoptosis in skeletal muscle, also contribute to insulin resistance, moreover, inhibiting caspase-3 reversed insulin resistance [44–46].
The enrichment pathways have been associated with insulin signal transduction, inflammation, cancer, hypoxia, cellular proliferation, differentiation and apoptosis, and energy metabolism. Dysfunction of the insulin receptor and signal transduction should be a crucial contributor [47]. Insulin signaling pathways activated by the insulin binding to the insulin receptor [48], induced insulin receptor auto-phosphorylation and then recruited insulin receptor substrates [49]. Signaling molecules downstream of insulin receptor substrates activated via phosphorylation by different kinases, such as PI3K and AKT, mTOR, ERK, FOXO, AMPK, and GSK3 [50]. The defect of insulin signaling pathways results in abnormal glucose homeostasis and ensued insulin resistance [47].
Of the top 10 node genes, INS, AKT1, and MAPK3 were proximal insulin receptor substrates, and played a key role in insulin signal transduction [47]. IL-6 and TNF were inflammatory mediators and always featured as candidates promoting insulin resistance [41]. VEGFA was related to hypoxia [43]. TP53 induced apoptosis and contributed to insulin resistance [45]. EGF was reported that played an insulin-like role in glucose transport and lipolysis [51]. EGF binding to EGF receptor (EGFR) increased the tyrosine phosphorylation and augmented the insulin-mediated downstream signaling in the insulin resistance state [51,52]. Src participated in cellular processes such as gene transcription, cell adhesion, and apoptosis [53]. It regulates inflammation-mediated metabolic disorders by phosphorylating peroxisome proliferator-activated receptors γ (PPAR-γ) and inhibiting pro-inflammatory genes in adipose tissue [54]. Another study reported that saturated fatty acids activated Jun N terminal kinase (JNK) and induced insulin resistance by altering the membrane distribution of c-Src [55].
Several limitations of this study should be addressed. The IRRGs were collected from publications. The criterion of IRRGs included in the 4 databases varied. And the heterogeneity of the validated methods in original researches should be considered. The roles of genes required to further verify that a gene caused insulin resistance or gene expression was altered by insulin resistance. Moreover, the insulin resistance occurred in different tissues and the mechanism related to insulin resistance may be different. However, the mechanism disclosed in the present study was an overview without tissue independent. In addition, all the included molecules had been previously validated and reported, and no outstanding findings of individual genes. However, it was a novel model of bioinformatics analysis based on validated molecules, it provided solid evidence for the regulatory network of insulin resistance.
Conclusions
The present study used a landscape about the IRRGs from disease-gene databases and illustrated the most possible underlying mechanism of insulin resistance by bioinformatics analysis based on IRRGs. Our results strongly suggest that inflammatory response, apoptotic process, hypoxia, signal transduction, and homeostasis may significantly play to the development and progression of insulin resistance. Our find revealed knowledge of mechanisms of insulin resistance and provided potential molecules for outcome prediction and novel targets for treatment.
Figures
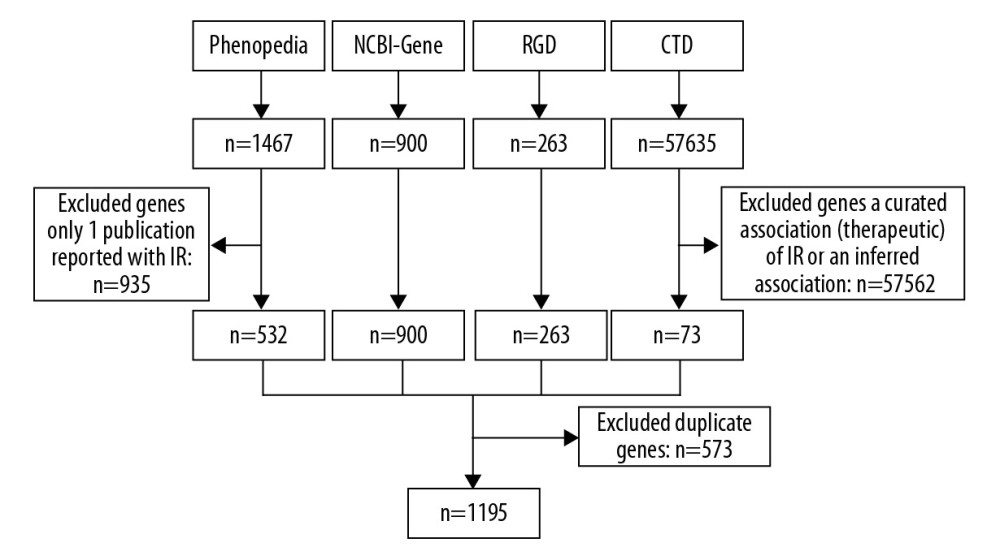 Figure 1. A flowchart of collecting IRRGs. IRRGs – insulin resistance-related genes.
Figure 1. A flowchart of collecting IRRGs. IRRGs – insulin resistance-related genes. 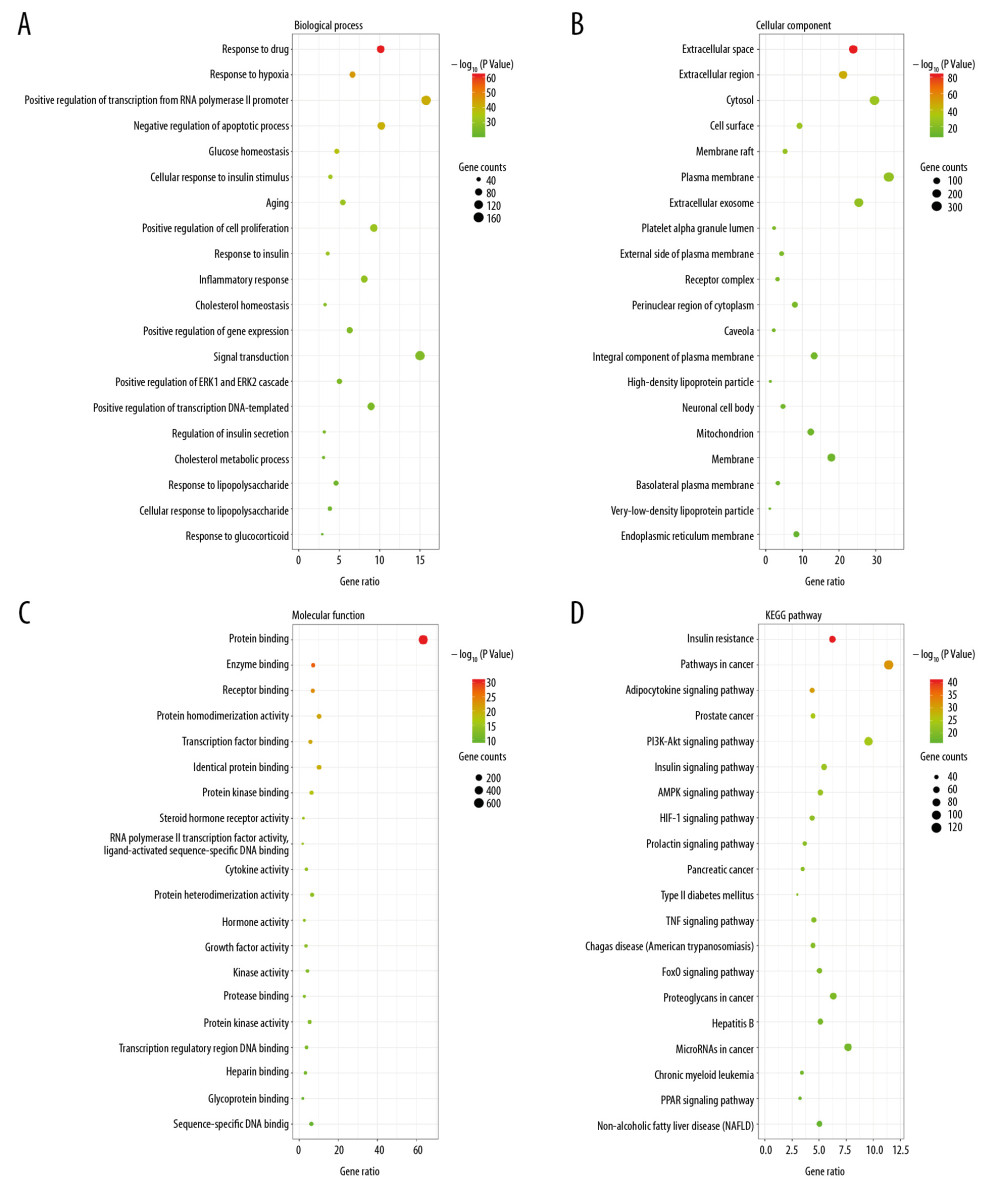 Figure 2. The GO and KEGG pathway analysis of IRRGs in DAVID. The top 20 terms were displayed. (A) Biological process. (B) Cellular component. (C) Molecular function. (D) KEGG pathway. GO – Gene Ontology; KEGG – Kyoto Encyclopedia of Genes and Genomes; IRRGs – insulin resistance-related genes; DAVID – Database for Annotation, Visualization and Integrated Discovery.
Figure 2. The GO and KEGG pathway analysis of IRRGs in DAVID. The top 20 terms were displayed. (A) Biological process. (B) Cellular component. (C) Molecular function. (D) KEGG pathway. GO – Gene Ontology; KEGG – Kyoto Encyclopedia of Genes and Genomes; IRRGs – insulin resistance-related genes; DAVID – Database for Annotation, Visualization and Integrated Discovery.  Figure 3. The top 4 modules PPI networks and responding biological process. PPI – protein–protein interaction.
Figure 3. The top 4 modules PPI networks and responding biological process. PPI – protein–protein interaction. 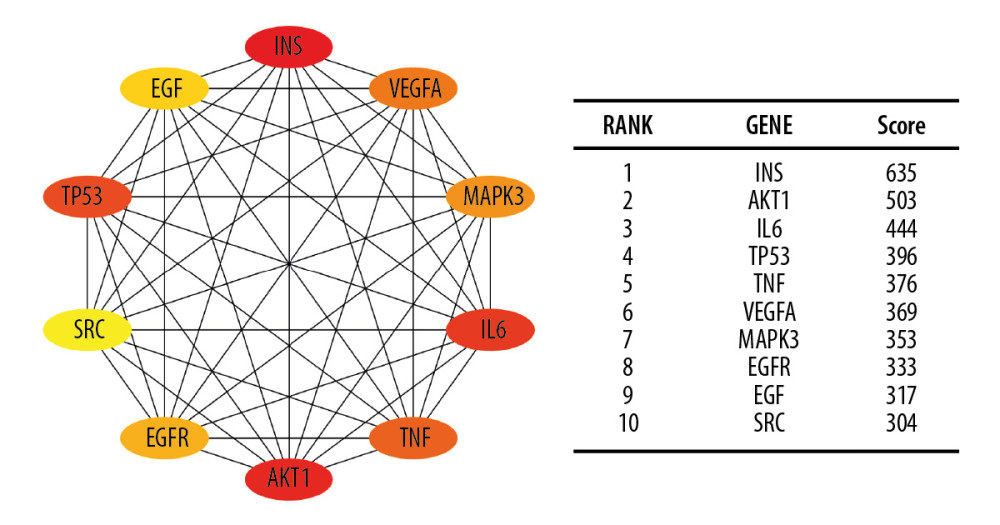 Figure 4. The top 10 hub genes of IRRGs. The more forward ranking was indicated by a redder color. IRRGs – insulin resistance-related genes.
Figure 4. The top 10 hub genes of IRRGs. The more forward ranking was indicated by a redder color. IRRGs – insulin resistance-related genes. References
1. Samuel VT, Shulman GI, The pathogenesis of insulin resistance: Integrating signaling pathways and substrate flux: J Clin Invest, 2016; 126; 12-22
2. Goldberg IJ, Diabetic dyslipidemia: Causes and consequences: J Clin Endocrinol Metab, 2001; 86; 965-71
3. Zhou MS, Schulman IH, Zeng Q, Link between the renin-angiotensin system and insulin resistance: Implications for cardiovascular disease: Vasc Med, 2012; 17; 330-41
4. Kim JA, Montagnani M, Koh KK, Quon MJ, Reciprocal relationships between insulin resistance and endothelial dysfunction: Molecular and pathophysiological mechanisms: Circulation, 2006; 113; 1888-904
5. Hirosumi J, Tuncman G, Chang L, A central role for JNK in obesity and insulin resistance: Nature, 2002; 420; 333-36
6. Tenenbaum A, Adler Y, Boyko V, Insulin resistance is associated with increased risk of major cardiovascular events in patients with preexisting coronary artery disease: Am Heart J, 2007; 153; 559-65
7. Howard G, O’Leary DH, Zaccaro D, Insulin sensitivity and atherosclerosis. The Insulin Resistance Atherosclerosis Study (IRAS) Investigators: Circulation, 1996; 93; 1809-17
8. Uruska A, Araszkiewicz A, Zozulinska-Ziolkiewicz D, Insulin resistance is associated with microangiopathy in type 1 diabetic patients treated with intensive insulin therapy from the onset of disease: Exp Clin Endocrinol Diabetes, 2010; 118; 478-84
9. de la Monte SM, Insulin resistance and neurodegeneration: Progress towards the development of new therapeutics for Alzheimer’s disease: Drugs, 2017; 77; 47-65
10. Izquierdo AG, Crujeiras AB, Role of epigenomic mechanisms in the onset and management of insulin resistance: Rev Endocr Metab Disord, 2019; 20; 89-102
11. Bluher M, Adipose tissue inflammation: A cause or consequence of obesity-related insulin resistance?: Clin Sci (Lond), 2016; 130; 1603-14
12. Houstis N, Rosen ED, Lander ES, Reactive oxygen species have a causal role in multiple forms of insulin resistance: Nature, 2006; 440; 944-48
13. Cheng Z, Tseng Y, White MF, Insulin signaling meets mitochondria in metabolism: Trends Endocrinol Metab, 2010; 21; 589-98
14. Flamment M, Hajduch E, Ferre P, Foufelle F, New insights into ER stress-induced insulin resistance: Trends Endocrinol Metab, 2012; 23; 381-90
15. Yip S-C, Saha S, Chernoff J, PTP1B: A double agent in metabolism and oncogenesis: Trends Biochem Sci, 2010; 35; 442-49
16. Piya MK, McTernan PG, Kumar S, Adipokine inflammation and insulin resistance: The role of glucose, lipids and endotoxin: J Endocrinol, 2013; 216; T1-15
17. Moraes-Vieira PM, Saghatelian A, Kahn BB: Diabetes, 2016; 65; 1808-15
18. Zhang Z, Liu H, Liu J, Akt activation: A potential strategy to ameliorate insulin resistance: Diabetes Res Clin Pract, 2019; 156; 107092
19. Ozaki K-i, Awazu M, Tamiya M, Targeting the ERK signaling pathway as a potential treatment for insulin resistance and type 2 diabetes: Am J Physiol Endocrinol Metab, 2016; 310; E651
20. Hosogai N, Fukuhara A, Oshima K, Adipose tissue hypoxia in obesity and its impact on adipocytokine dysregulation: Diabetes, 2007; 56; 901-11
21. Sun G, Li Y, Peng Y, Identification of differentially expressed genes and biological characteristics of colorectal cancer by integrated bioinformatics analysis: J Cell Physiol, 2019; 234; 15215-24
22. Yang Y, Wang Y, Zhou K, Hong A, Constructing regulatory networks to identify biomarkers for insulin resistance: Gene, 2014; 539; 68-74
23. Zhang Y, Zheng Y, Fu Y, Wang C, Identification of biomarkers, pathways and potential therapeutic agents for white adipocyte insulin resistance using bioinformatics analysis: Adipocyte, 2019; 8; 318-29
24. Morey JS, Ryan JC, Van Dolah FM, Microarray validation: factors influencing correlation between oligonucleotide microarrays and real-time PCR: Biol Proced Online, 2006; 8; 175-93
25. Brown GR, Hem V, Katz KS, Gene: A gene-centered information resource at NCBI: Nucleic Acids Res, 2015; 43; D36-42
26. Davis AP, Grondin CJ, Johnson RJ, The Comparative Toxicogenomics Database: Update 2019: Nucleic Acids Res, 2019; 47; D948-54
27. Shimoyama M, De Pons J, Hayman GT, The Rat Genome Database 2015: Genomic, phenotypic and environmental variations and disease: Nucleic Acids Res, 2015; 43; D743-50
28. Yu W, Clyne M, Khoury MJ, Gwinn M, Phenopedia and genopedia: Disease-centered and gene-centered views of the evolving knowledge of human genetic associations: Bioinformatics, 2010; 26; 145-46
29. : The National Center for Biotechnology Information’s Gene database www.ncbi.nlm.nih.gov/gene
30. : The Comparative Toxicogenomics Database http://ctdbase.org/
31. : The Rat Genome Database http://ctdbase.org/
32. : Phenopedia https://phgkb.cdc.gov/PHGKB/startPagePhenoPedia.action
33. , Database for Annotation, Visualization and Integrated Discovery (DAVID): Bioinformatics Resources 6.8 https://david.ncifcrf.gov/home.jsp
34. Huang DW, Sherman BT, Tan Q, DAVID Bioinformatics Resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists: Nucleic Acids Res, 2007; 35; W169-75
35. Szklarczyk D, Morris JH, Cook H, The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible: Nucleic Acids Res, 2017; 45; D362-68
36. : The Search Tool for the Retrieval of Interacting Genes database http://string-db.org/
37. Saiselet M, Floor S, Tarabichi M, Thyroid cancer cell lines: An overview: Front Endocrinol (Lausanne), 2012; 3; 133
38. Chin CH, Chen SH, Wu HH, CytoHubba: Identifying hub objects and sub-networks from complex interactome: BMC Syst Biol, 2014; 8(Suppl 4); S11
39. Trayhurn P, Wood IS, Adipokines: Inflammation and the pleiotropic role of white adipose tissue: Br J Nutr, 2007; 92; 347-55
40. Ye J, Gao Z, Yin J, He Q, Hypoxia is a potential risk factor for chronic inflammation and adiponectin reduction in adipose tissue of OB/OB and dietary obese mice: Am J Physiol Endocrinol Metab, 2007; 293; E1118-28
41. Dandona P, Aljada A, Bandyopadhyay A, Inflammation: The link between insulin resistance, obesity and diabetes: Trends Immunol, 2004; 25; 4-7
42. Rasouli N, Adipose tissue hypoxia and insulin resistance: J Investig Med, 2016; 64; 830-32
43. Yin J, Gao Z, He Q, Role of hypoxia in obesity-induced disorders of glucose and lipid metabolism in adipose tissue: Am J Physiol Endocrinol Metab, 2009; 296; E333-42
44. Alkhouri N, Gornicka A, Berk MP, Adipocyte apoptosis, a link between obesity, insulin resistance, and hepatic steatosis: J Biol Chem, 2010; 285; 3428-38
45. Derdak Z, Lang CH, Villegas KA, Activation of p53 enhances apoptosis and insulin resistance in a rat model of alcoholic liver disease: J Hepatol, 2011; 54; 164-72
46. Turpin SM, Lancaster GI, Darby I, Apoptosis in skeletal muscle myotubes is induced by ceramides and is positively related to insulin resistance: Am J Physiol Endocrinol Metab, 2006; 291; E1341-50
47. Petersen MC, Shulman GI, Mechanisms of insulin action and insulin resistance: Physiol Rev, 2018; 98; 2133-23
48. Boucher J, Kleinridders A, Kahn CR, Insulin receptor signaling in normal and insulin-resistant states: Cold Spring Harb Perspect Biol, 2014; 6; a009191
49. Haeusler RA, McGraw TE, Accili D, Biochemical and cellular properties of insulin receptor signalling: Nat Rev Mol Cell Biol, 2018; 19; 31-44
50. Copps KD, White MF, Regulation of insulin sensitivity by serine/threonine phosphorylation of insulin receptor substrate proteins IRS1 and IRS2: Diabetologia, 2012; 55; 2565-82
51. Gogg S, Smith U, Epidermal growth factor and transforming growth factor α mimic the effects of insulin in human fat cells and augment downstream signaling in insulin resistance: J Biol Chem, 2002; 277; 36045-51
52. Borisov N, Aksamitiene E, Kiyatkin A, Systems-level interactions between insulin-EGF networks amplify mitogenic signaling: Mol Syst Biol, 2009; 5; 256
53. Pan Y, Li G, Zhong H, RIG-I inhibits pancreatic β cell proliferation through competitive binding of activated Src: Sci Rep, 2016; 6; 28914
54. Choi S, Jung JE, Yang YR, Novel phosphorylation of PPARgamma ameliorates obesity-induced adipose tissue inflammation and improves insulin sensitivity: Cell Signal, 2015; 27; 2488-95
55. Holzer RG, Park EJ, Li N, Saturated fatty acids induce c-Src clustering within membrane subdomains, leading to JNK activation: Cell, 2011; 147; 173-84
Figures
 Figure 1. A flowchart of collecting IRRGs. IRRGs – insulin resistance-related genes.
Figure 1. A flowchart of collecting IRRGs. IRRGs – insulin resistance-related genes. Figure 2. The GO and KEGG pathway analysis of IRRGs in DAVID. The top 20 terms were displayed. (A) Biological process. (B) Cellular component. (C) Molecular function. (D) KEGG pathway. GO – Gene Ontology; KEGG – Kyoto Encyclopedia of Genes and Genomes; IRRGs – insulin resistance-related genes; DAVID – Database for Annotation, Visualization and Integrated Discovery.
Figure 2. The GO and KEGG pathway analysis of IRRGs in DAVID. The top 20 terms were displayed. (A) Biological process. (B) Cellular component. (C) Molecular function. (D) KEGG pathway. GO – Gene Ontology; KEGG – Kyoto Encyclopedia of Genes and Genomes; IRRGs – insulin resistance-related genes; DAVID – Database for Annotation, Visualization and Integrated Discovery. Figure 3. The top 4 modules PPI networks and responding biological process. PPI – protein–protein interaction.
Figure 3. The top 4 modules PPI networks and responding biological process. PPI – protein–protein interaction. Figure 4. The top 10 hub genes of IRRGs. The more forward ranking was indicated by a redder color. IRRGs – insulin resistance-related genes.
Figure 4. The top 10 hub genes of IRRGs. The more forward ranking was indicated by a redder color. IRRGs – insulin resistance-related genes. In Press
05 Mar 2024 : Clinical Research
Role of Critical Shoulder Angle in Degenerative Type Rotator Cuff Tears: A Turkish Cohort StudyMed Sci Monit In Press; DOI: 10.12659/MSM.943703
06 Mar 2024 : Clinical Research
Comparison of Outcomes between Single-Level and Double-Level Corpectomy in Thoracolumbar Reconstruction: A ...Med Sci Monit In Press; DOI: 10.12659/MSM.943797
21 Mar 2024 : Meta-Analysis
Economic Evaluation of COVID-19 Screening Tests and Surveillance Strategies in Low-Income, Middle-Income, a...Med Sci Monit In Press; DOI: 10.12659/MSM.943863
10 Apr 2024 : Clinical Research
Predicting Acute Cardiovascular Complications in COVID-19: Insights from a Specialized Cardiac Referral Dep...Med Sci Monit In Press; DOI: 10.12659/MSM.942612
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952