02 August 2020: Clinical Research
Identification of Differentially Expressed Proteins in Serum of Obese Patients by Isobaric Tags for Relative and Absolute Quantification (iTRAQ)-Coupled 2D LC-MS
Ying Zhang1ABCDEF, Zhong Chen2BCDF, Yue He2BCDF, Jianxia Wang2CDF, Minhui Jiang2BDF, Jianjuan Xu2CF, Minghua Chen2CF, Yaling Feng2CG*DOI: 10.12659/MSM.924882
Med Sci Monit 2020; 26:e924882
Abstract
BACKGROUND: The aim of this study was to identify the differentially expressed proteins of obese patients compared with normal participants and to provide a potential target for future investigation of obesity.
MATERIAL AND METHODS: We enrolled 10 obese male adults and 10 matched normal subjects. Serum samples were collected to get total protein extraction, denaturation, deoxidation, and enzymatic hydrolysis. Differentially expressed proteins were distinguished with mass spectrometry after samples were labeled with iTRAQ.
RESULTS: A total of 9622 differentially expressed peptides were identified, corresponding to 733 proteins; 118 proteins of these showed significant differential expression, with 15 upregulated and 103 downregulated.
CONCLUSIONS: iTRAQ is an effective technique to identify differentially expressed proteins in obese patients. The development of obesity is correlated with a series of complex elements and mutual effects. The proteins identified in this study may provide novel directions and targets for future pathological studies of obesity.
Keywords: 14-3-3 Proteins, Obesity, Abdominal, Serum Albumin, Bovine, Blood Proteins, Body Mass Index, Case-Control Studies, Chromatography, Liquid, Gene Expression Profiling, Gene Expression Regulation, gene ontology, Metabolic Networks and Pathways, Molecular Sequence Annotation, Staining and Labeling, tandem mass spectrometry
Background
Obesity refers to excessive accumulation and abnormal distribution of fat in the body, usually accompanied by weight gain. The World Health Organization (WHO) defines obesity as an abnormal or excessive accumulation of fat that can lead to health damage [1]. According to the survey of nutrition and health status of Chinese residents of 2002, the overweight rate and obesity rate of Chinese residents were 17.6% and 5.6% respectively. The sum of the 2 was 23.2%, which was close to a quarter of the total population. Obesity has become an important disease affecting the health of Chinese residents [2]. Obesity is a major risk factor for many diseases, such as hypertension, diabetes, dyslipidemia, coronary heart disease, myocardial infarction, stroke, and several types of cancer, including breast cancer. It has been recognized as the fifth most important major risk factor affecting health [3]. Some studies had found that ceruloplasmin, fibrinogen, and apolipoprotein A-IV were overexpressed in plasma of obese people; therefore, they might be potential protein markers of obesity [4,5]. In contrast, apolipoprotein A-1 and apolipoprotein E were lower in plasma of obese and overweight people compared with the normal population [5]. Some studies demonstrated that level of the thermogenic enzymes was higher while the level of lipid synthase was lower in obesity-resistant mice [6]. Cytochrome b-cl complex subunit 1, enol3, monoglyceride lipase, and glucose-6 phosphate dehydrogenase 1 participate in energy metabolism in mice, such as glycolysis and visceral adipose tissue formation; thus, they may also be involved in the molecular mechanism and metabolic pathway of obesity [7].
Proteomics is a recently-developed method to study the molecular mechanism of diseases. iTRAQ technology is a proteomic technology introduced by Applied Biosystems, Inc. in 2004, in which 4 or 8 samples under the same experimental conditions can be quantitatively studied simultaneously by using isotope labeling technology. iTRAQ reagents comprised heterotopic labeling molecules with molecular weight of 145 Da. The labeling molecules are made up of reporting group with molecular weights of 114, 115, 116, 11, 118, 119, and 121Da, equilibrium group, and a same polypeptide reaction group. The technique has very good accuracy and high repeatability. It can be used to analyze several different samples simultaneously. When combined with the chromatographic separation system, it can be used to detect large-scale protein alterations by detecting microgram-level amounts of protein, thus providing large-scale quantitative information on proteins. Another advantage is its wide application, as it can assess any type of protein. Compared with gel electrophoresis, this technology can be used to identify large molecular weight proteins, extremely acidic proteins, and extremely basic proteins, as well as low solubility and even insoluble membrane proteins; thus, the range of proteins identified is greatly expanded [8].
In the present study, we used iTRAQ technique to identify the differentially expressed proteins of obese patients compared with normal participants, to explore the potential molecular mechanism of obesity.
Material and Methods
PARTICIPANTS AND ENROLLMENT:
Adults with BMI >28.0 kg/m2 were categorized as obese, as specified in the Guidelines for the Prevention and Control of Overweight and Obesity in Adults of China promulgated and implemented by the Ministry of Health in 2002 [9]. We included 10 obese subjects and 10 normal subjects. Inclusion criteria of the obesity group were: male; age 18–44 years old; BMI ≥28.0 kg/m2; no organic diseases; and normal blood pressure and serum glucose (Table 1). Matched controls were enrolled in a ratio of 1: 1. The inclusion criteria of the control group were: male; age 18–44 years old; 18.5 kg/m2 ≤BMI <24.0 kg/m2; no organic diseases; and normal blood pressure, serum glucose, serum lipid, and hemoglobin levels. All subjects signed the informed consent. The research was approved by the Ethics Committee of Wuxi Maternal and Child Health Hospital Affiliated to Nanjing Medical University.
REAGENTS AND INSTRUMENT:
The system in the MARS.14 chromatographic column was obtained from Aglient Co.; the iTRAQ kit, QSTAR XL mass spectrometer, and Protein Pilot4.2 software were obtained from Applied Biosystems (USA); the liquid chromatograph (20AD) was obtained from Shimadzu Co. (Japan); Strong Cation Exchange Chromatography (2.1×100 mm, 5 um, 300 A) was obtained for the Nest Group Inc. System (USA); and the ZORBAX 300SB-C18 column (5 um, 300A, 0.1×150 mm) was obtained from Microm Co. (USA).
SAMPLE COLLECTION AND PREPARATION:
We collected 4 mL fasting venous blood and placed it in a tube. After being frozen on ice for 30 min, the sample was centrifuged under 2000 r/min for 10 min at 4°C. Supernatant was collected in 1.5-mL EP tubes before being stored at −80°C.
REMOVAL OF HIGH-ABUNDANCE PROTEINS:
High-abundance proteins were removed using the multiple affinity removal system human 14 chromatograph. The fractions collected were concentrated by freeze-drying.
ITRAQ LABELING:
One hundred ug samples of each group were added with reducing reagent, at 60°C for 1 h. Then, cysteine blocking reagent was added and placed at room temperature for 10 min. Trypsin was added and enzymatic hydrolysis was allowed at 37°C overnight. Then, samples were centrifuged to deposit, and iTRAQ labeling reagent was added to each sample. Reaction was allowed for 1 h at room temperature and 3 times the volume of super-pure water was added to dilute the labeling reagent. The sample of the obese group was labeled with iTRAQ 118 and samples of the normal group were labeled with iTRAQ 117. The labeled samples were mixed in a low-adsorption tube and underwent vacuum centrifugal evaporation drying, followed by storage at −80°C.
STRONG CATION EXCHANGE CHROMATOGRAPHY (SCX) AND MASS SPECTROMETRIC ANALYSIS:
We added 200 uL SCX buffer A to the labeled sample mixture, stratified by SCX. After stratification, samples were analyzed by nano liquid chromatography electron spray ionization mass spectrometry (nano-LC-ESI-MS/MS) system. The scanning range of paper and pen was 400–1800 m/z.
MASS SPECTROMETRIC DATA ANALYSIS:
The data obtained with mass spectrometric analysis were retrieved by Protein Pilot 4.2 software, using the Paragon method. Protein identification confidence level was set as 95%, Prot Score >1.3, and a peptide above 95% matched peptides in the library. At the same time, a protein reverse library search was made. The FDR <5%, Ratio >1.2 or Ratio <0.8 was regarded as differential expression.
GENE ONTOLOGY (GO) FUNCTIONAL ANALYSIS:
All differentially expressed proteins identified by the approaches above were assigned their gene symbol via the PANTHER database (Protein Analysis through Evolutionary Relationships,
DATA ANALYSIS AND STATISTICS:
The data are presented as mean±SD and were assessed by
Results
AGE, BMI, BLOOD PRESSURE, AND GLUCOSE OF SUBJECTS:
The BMI of obese group was significantly higher than that of the control group (Table 1). However, their age, systolic blood pressure, diastolic blood pressure, and glucose were normal and had no significant differences (p>0.05) (Table 1).
BASIC DATA OF IDENTIFIED PROTEINS:
A total of 9622 differential peptides were identified, corresponding to 733 proteins; 118 proteins showed significant differential expression, with 15 upregulated and 103 downregulated (Table 2). The upregulated proteins included IGL@ protein, C-reactive protein, Putative uncharacterized protein, and Suprabasin (Table 3). The downregulated proteins included Thioredoxin reductase 2, Costars family protein ABRACL, Kelch domain containing 5, and POTE ankyrin domain family member J (Table 4). The downregulated Apo-lipoproteins included apolipoprotein B variant, ApoB, and ApoM. The downregulated energy metabolism-related proteins included phosphoglycerate kinase 1, N-fatty-acyl-amino acid synthase, and glucose-6-phosphate isomerase.
ENRICHMENT ANALYSIS OF DIFFERENTIALLY EXPRESSED PROTEINS:
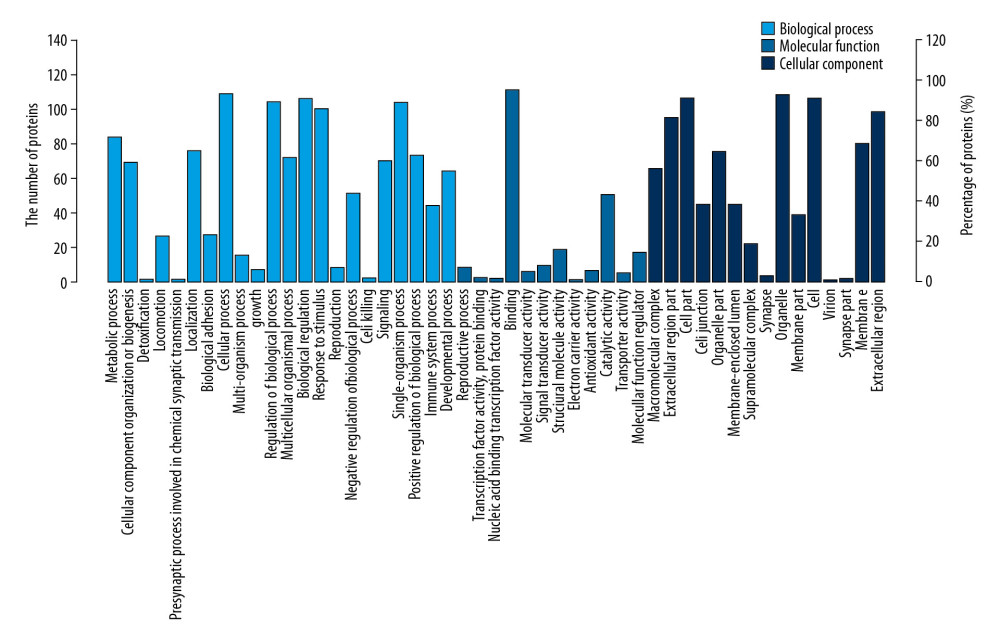
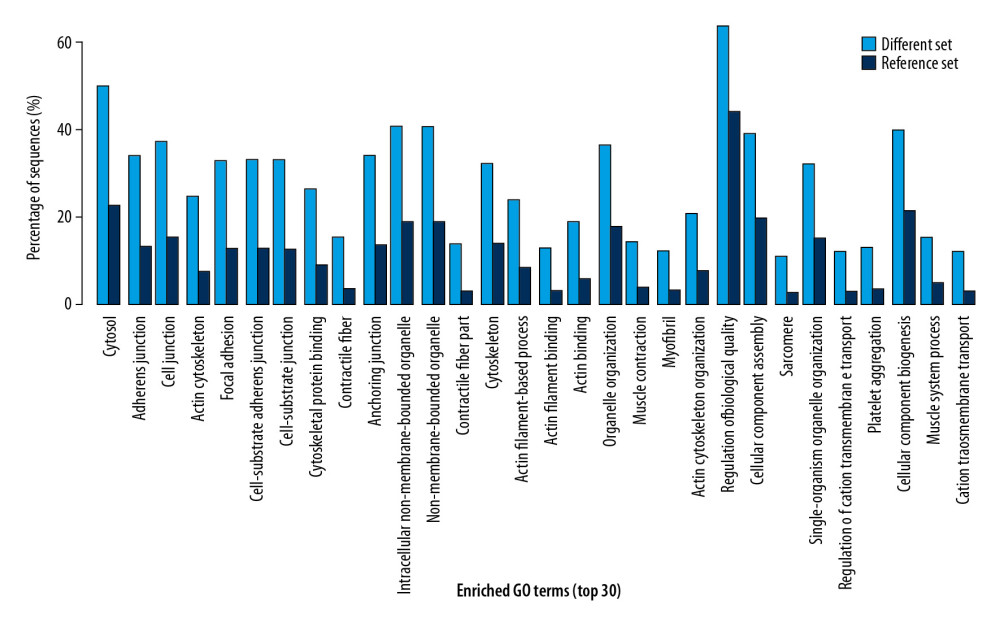
The results of Go functional annotation analysis indicated the percentage of differential expression of proteins involved in every biological procedure, molecular function, and cellular component (Figure 1). The items with high percentage of differential proteins were involved in included cellular processes, signaling process, binding, cell part, and organelles (Figure 1). Furthermore, the top 30 enriched GO terms indicated the significantly enriched GO terms involved by the differentially expression of proteins (Figure 2). The significantly enriched GO terms included actin cytoskeleton, contractile fiber, myofibril, and platelet aggregation (Figure 2).
KEGG ENRICHMENT PATHWAY ANALYSIS OF DIFFERENTIALLY EXPRESSED PROTEINS:
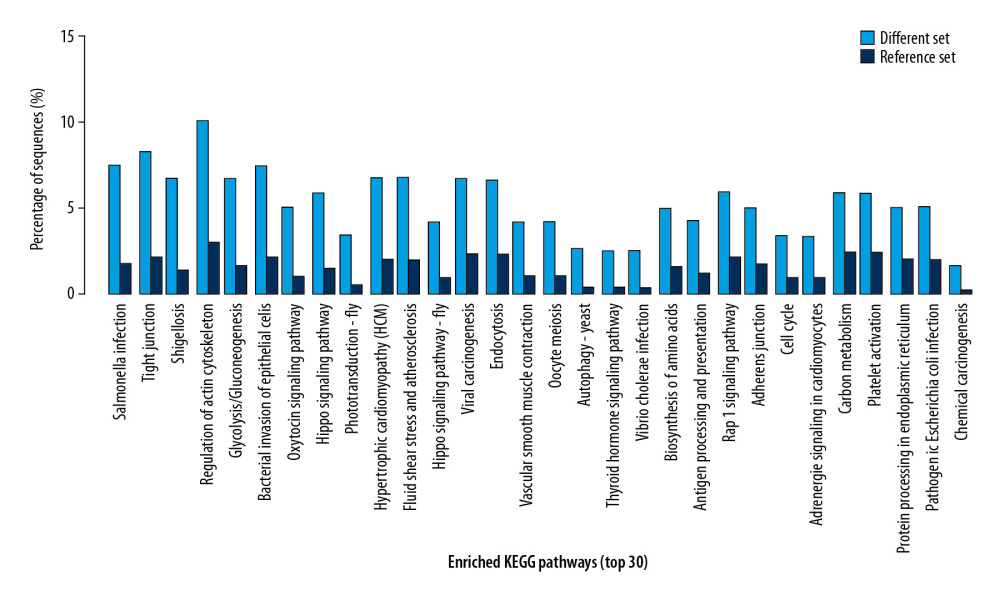
The top 30 enriched pathways involved with differential proteins were demonstrated. These pathways included salmonella infection, tight junction, shigellosis, and regulation of actin cytoskeleton (Figure 3).
Discussion
The incidence of obesity in China is increasing rapidly. At present, more than one-half of males are overweight. The increase of abdominal obesity in men is fairly obvious, even in young males. It is necessary to investigate obesity to prevent and control the development and progression of obesity. In this study, we used iTRAQ technology to analyze the serum proteomics among obese male patients, and identified several obesity-related proteins, thereby providing a basis for further research on obesity.
According to the results of this study, the level of C-reactive protein was significantly higher in serum of obese patients, which was consistent with previous studies [10,11]. We found that upregulation of C-reactive protein promoted chronic inflammatory status. C-reactive protein is a pro-inflammation factor and can strongly predict the risk of cardiovascular events, since it can mediate cardiovascular impairment. Some studies have demonstrated that C-reactive protein in adipose cells can inhibit expression and secretion of adiponectin [10,11]. Adiponectin is a specific protein generated by adipose cells and is associated with glucose utilization, fatty acid oxidation, and energy consumption, thus playing an important role in obesity [12]. In addition, adiponectin is importantly involved in formation of arteriosclerosis plaque. It can inhibit inflammation reaction through various mechanisms, thereby preventing arteriosclerosis formation. However, more investigations are needed to explore the reverse association between C-reactive protein and adiponectin and its impact on obesity.
We found that the levels of apolipoprotein B variant, ApoB, and ApoM were significantly lower in obese patients. Relevant studies have demonstrated that ApoA1, Apo A-IV, and ApoB levels were downregulated and ApoC-III was upregulated in serum of obese patients. The ratio of ApoB/ApoA1 is tightly associated with obesity, which is consistent with that of previous studies [13–16]. Therefore, ApoB may be a potential marker of obesity. ApoM is a recently-discovered apolipoprotein that is a member of the lipoprotein superfamily. It mainly exists in high-density lipoprotein particles. ApoM is involved in pre-beta-HDL synthesis, promotes vascular protection function of HDL, and promotes insulin release [17]. It is regarded as being potentially protective against atherosclerosis, coronary disease, and diabetes [18,19]. Similar to C-reactive protein, recent studies have found that ApoM expression was significantly regulated in some inflammation-related diseases such as hepatitis, sepsis, pneumonia, and arthritis [20]. Thus, apart from relying on the lipid transportation function of ApoM, the association between ApoM and obesity might be more complicated, and further investigations are needed to elucidate the mechanism involved.
Moreover, levels of phosphoglycerate kinase 1, N-fatty-acyl-amino acid synthase and glucose-6-phosphate isomerase, which are closely associated with energy metabolism, were significantly lower in obese patients. However, the levels of adiponectin and leptin were not assessed, partly due to the upregulation of C-reactive protein. Therefore, studies with larger sample sizes are needed in the future.
Conclusions
The development of obesity may involve a series of complex elements and mutual effects. Using the high-volume iTRAQ technique, we identified several proteins which were obviously regulated in serum of obese subjects. These proteins may provide novel directions and targets for future pathological studies of obesity.
Tables
Table 1. Basic information of 20 participants (SBP – systolic blood pressure; DBP – diastolic blood pressure; Mean±SD).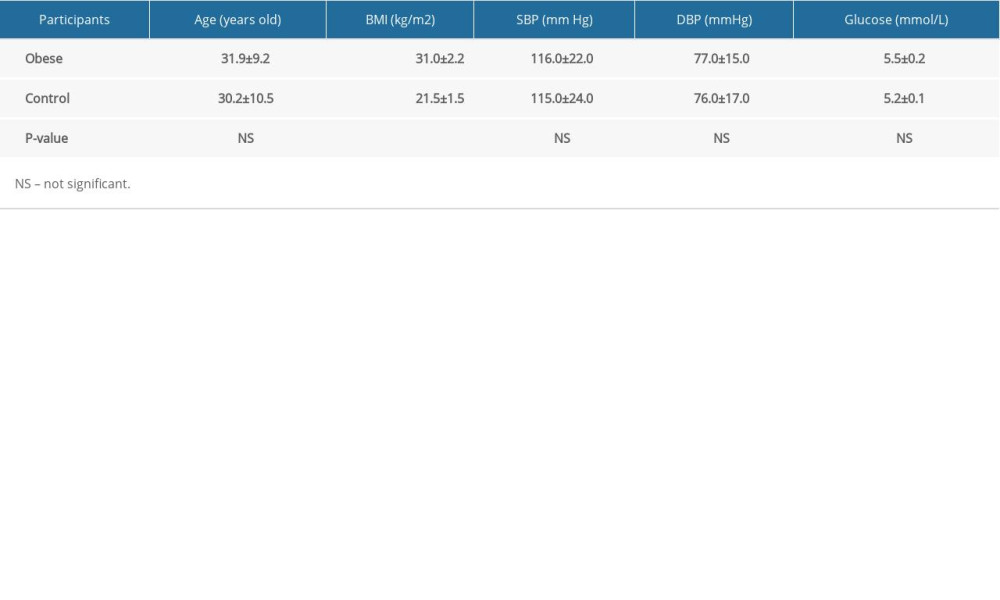 Table 2. General information of identified proteins.
Table 2. General information of identified proteins.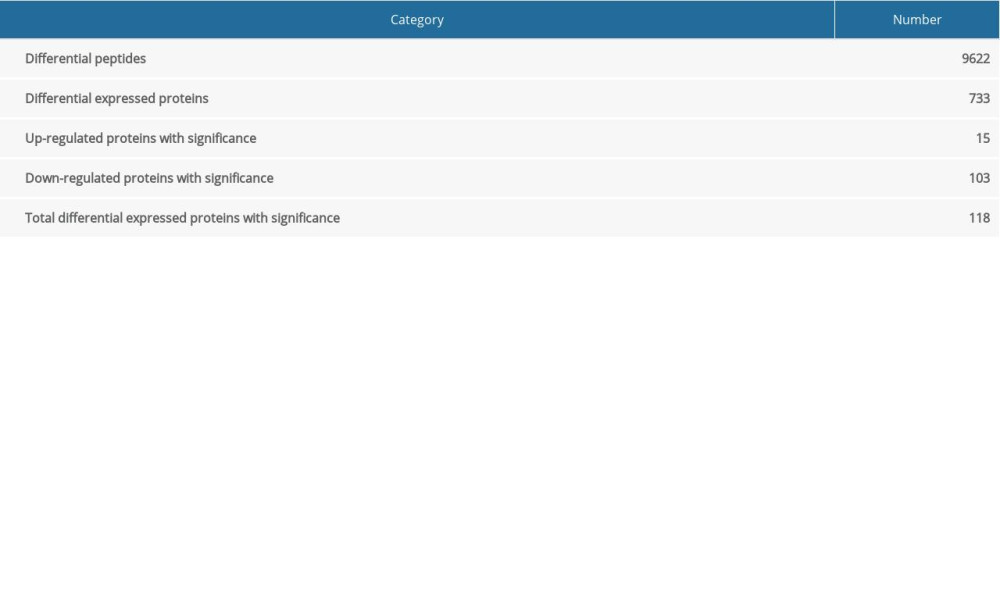 Table 3. List of all 15 up-regulated proteins.
Table 3. List of all 15 up-regulated proteins.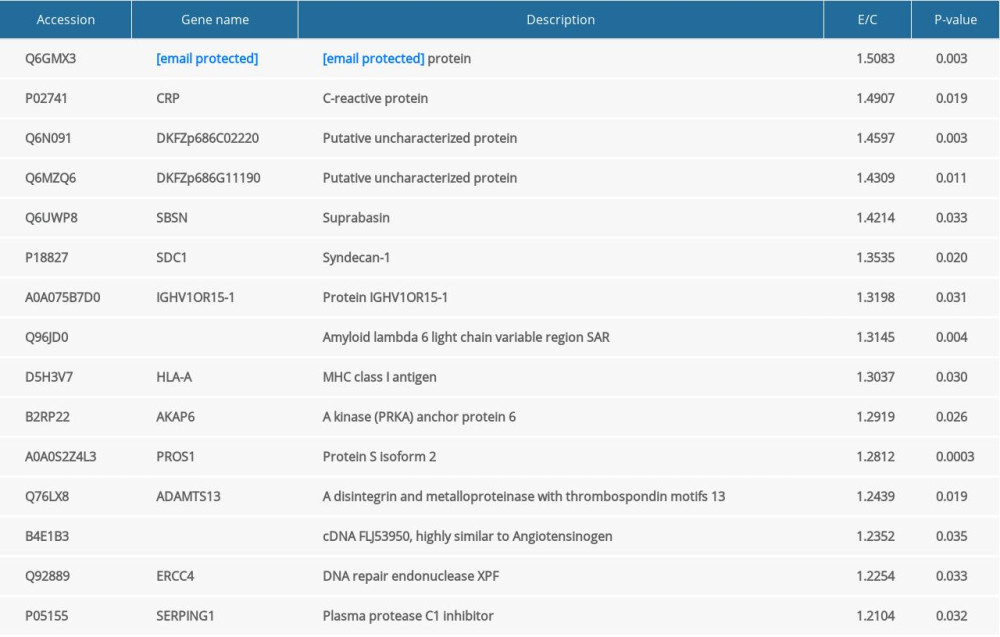 Table 4. Top 15 down-regulated proteins, Apo-lipoproteins and energy metabolism related proteins.
Table 4. Top 15 down-regulated proteins, Apo-lipoproteins and energy metabolism related proteins.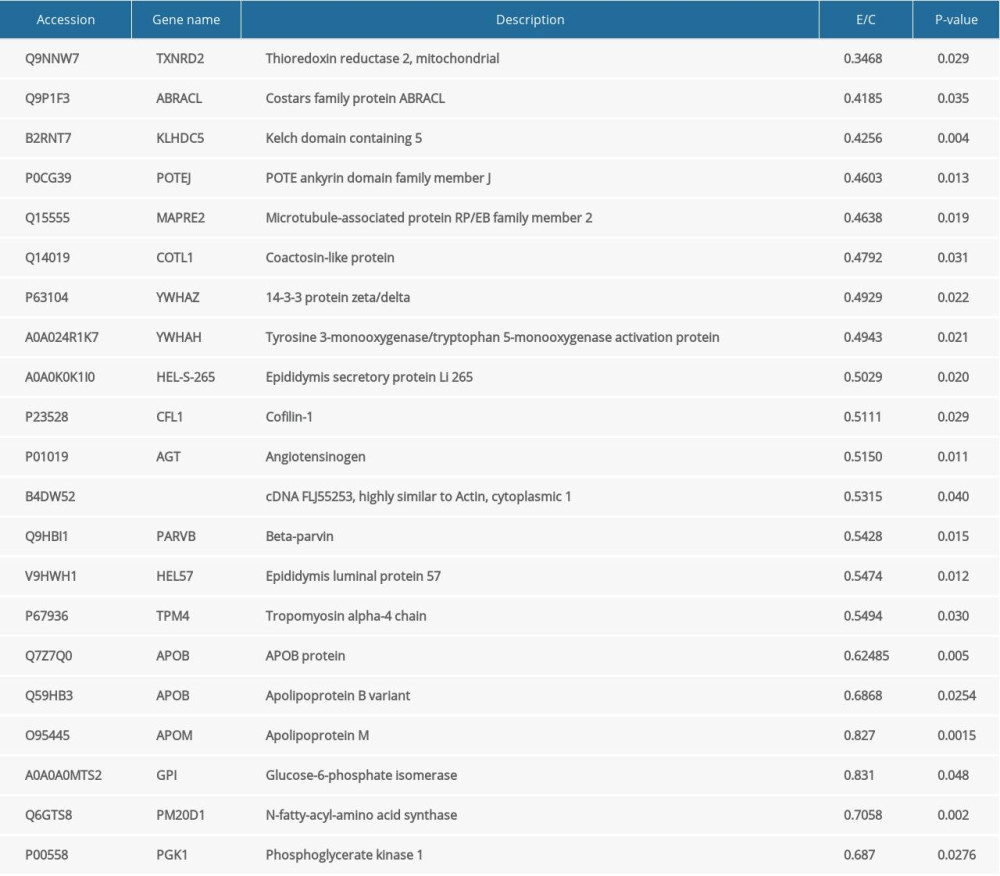
References
1. , Consensus of obesity prevention and treatment among Chinese adults: Chin J Endocrinol Metab, 2011; 27; 711-18
2. Wu Y, Ma G, Hu YThe current prevalence status of body overweight and obesity in China: Data from the China national nutrition and health survey: Zhonghua Yu Fang Yi Xue Za Zhi, 2005; 39(5); 316-20
3. Mackay J, Mensah GA, Mendis S: The atlas of heart disease and stroke, 2004; 5-6, Geneva, WHO
4. Kim OY, Shin MJ, Moon J, Plasma ceruloplasmin as a biomarker for obesity: A proteomic approach: Clin Biochem, 2011; 44(5–6); 351-56
5. Galata Z, Moschonis G, Makridakis M, Plasma proteomic analysis in obese and overweight prepubertal children: Eur J Clin Invest, 2011; 41(12); 1275-83
6. Joo JI, Oh TS, Kim DH, Differential expression of adipose tissue proteins between obesity-susceptible and resistant rats fed a high-fat diet: Proteomics, 2011; 11(8); 1429-48
7. Xie WD, Wang H, Zhang JF, Proteomic profile of visceral adipose tissue between low-fat diet-fed obesity-resistant and obesity-prone C57BL/6 mice: Mol Med Rep, 2010; 3(6); 1047-52
8. Wei C, Lai M, Mo X, Screening serum biomarkers in gastric cancer by iTRAQ-coupled 2D-LC-MS/MS: J Prac Med, 2014; 30; 1154-57
9. Li L, Rao K, Kong L, Survey on nutrition and health status of Chinese residents in 2002: Chin J Epidemiol, 2005; 26(7); 478-84
10. Wang J, Yu W, Xu JScreening of protein markers on the plasma of obese young men: Zhonghua Yu Fang Yi Xue Za Zhi, 2013; 47(2); 147-50
11. Gao P, Jiao Z, Ma J, Effects of C-reactive protein on regulation of adiponectin expression in human adipocytes: Journal of Harbin Medical University, 2011; 45; 27-29
12. Yamauchi T, Kamon J, Waki H, The fat-derived hormone adiponectin reverse insulin resistance associated with both lipotrophy and obesity: Nat Med, 2001; 7(8); 941-46
13. Li X, Hou Y, Wang Y, Analysis on levels of apolipoprotein, lipoprotein(a) and blood lipids in patients with adiposity: Journal of Tianjin Medical University, 2005; 11; 380-81
14. Zhang G, Wang Z, Huang H, Study on the clinical significance of overweight/obesity and diabetic patients serum apolipoprotein B and apolipoprotein A1 ratio: China Modern Doctor, 2013; 51; 54-56
15. Hui L, Gui-Fa XU, Gen-Sheng L, Relationship among Chinese serum apolipoprotein A-IV level, obesity and leptin level: Journal of Shangdong University, 2007; 45; 6-9
16. Suhong Wu, Bao-Jin C, Li-Wen Z, Study on the expression of Apo A5 in obese children and its correlation with adiponectin and fasting insulin: Journal of Clinical and Experimental Medicine, 2017; 16(5); 479-82
17. Ren K, Tang Z-L, Jiang Y, Apolipoprotein M: Clin Chim Acta, 2015; 446; 21-29
18. Ferrante M, Henckaerts L, Joossens M, New serological markers in inflammatory bowel disease are associated with complicated disease behavior: Gut, 2007; 56(10); 1394-403
19. Seow CH, Stempak JM, Xu W, Novel anti-glycan antibodies related to inflammatory bowel disease diagnosis and phenotype: Am J Gastroenterol, 2009; 104(6); 1426-34
20. Stephens NS, Stiffledeen J, Su X, Urinary NMR metabolomic profiles discriminate inflammatory bowel disease from healthy: J Crohns Colitis, 2013; 7(2); e42-48
Figures
Tables
 Table 1. Basic information of 20 participants (SBP – systolic blood pressure; DBP – diastolic blood pressure; Mean±SD).
Table 1. Basic information of 20 participants (SBP – systolic blood pressure; DBP – diastolic blood pressure; Mean±SD). Table 2. General information of identified proteins.
Table 2. General information of identified proteins. Table 3. List of all 15 up-regulated proteins.
Table 3. List of all 15 up-regulated proteins. Table 4. Top 15 down-regulated proteins, Apo-lipoproteins and energy metabolism related proteins.
Table 4. Top 15 down-regulated proteins, Apo-lipoproteins and energy metabolism related proteins. Table 1. Basic information of 20 participants (SBP – systolic blood pressure; DBP – diastolic blood pressure; Mean±SD).
Table 1. Basic information of 20 participants (SBP – systolic blood pressure; DBP – diastolic blood pressure; Mean±SD). Table 2. General information of identified proteins.
Table 2. General information of identified proteins. Table 3. List of all 15 up-regulated proteins.
Table 3. List of all 15 up-regulated proteins. Table 4. Top 15 down-regulated proteins, Apo-lipoproteins and energy metabolism related proteins.
Table 4. Top 15 down-regulated proteins, Apo-lipoproteins and energy metabolism related proteins. In Press
06 Mar 2024 : Clinical Research
Prevalence and Variation of Medical Comorbidities in Oral Surgery Patients: A Retrospective Study at Jazan ...Med Sci Monit In Press; DOI: 10.12659/MSM.943884
08 Mar 2024 : Clinical Research
Evaluation of Foot Structure in Preschool Children Based on Body MassMed Sci Monit In Press; DOI: 10.12659/MSM.943765
15 Apr 2024 : Laboratory Research
The Role of Copper-Induced M2 Macrophage Polarization in Protecting Cartilage Matrix in OsteoarthritisMed Sci Monit In Press; DOI: 10.12659/MSM.943738
07 Mar 2024 : Clinical Research
Knowledge of and Attitudes Toward Clinical Trials: A Questionnaire-Based Study of 179 Male Third- and Fourt...Med Sci Monit In Press; DOI: 10.12659/MSM.943468
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952