17 September 2020: Lab/In Vitro Research
Cyclin G2 Is Involved in the Proliferation of Placental Trophoblast Cells and Their Interactions with Endothelial Cells
Manni Sun1A, Shenghuan Liu1C, Jinlan Gao1C, Tao Meng2BC, Xuesha Xing1CD, Chen Chen1CD, Haiying Chen2E, Yang Luo1AB*DOI: 10.12659/MSM.926414
Med Sci Monit 2020; 26:e926414
Abstract
BACKGROUND: Remodeling of maternal spiral arteries after embryo implantation relies on well-regulated trophoblast functions. Although cyclin G2 (CCNG2) is thought to be involved in placental development and function, its role in trophoblasts and the mechanisms underlying placental development and function remain unclear. The present study investigated the potential role of CCNG2 in trophoblast cell proliferation and their interactions with endothelial cells.
MATERIAL AND METHODS: CCNG2 levels were modified by stable infection of HTR8/SVneo cells with lentiviruses overexpressing and silencing CCNG2. Cell proliferation was measured using CCK-8 assays. Network formation assays were performed using trophoblasts alone and co-cultured trophoblasts and endothelial cells to measure angiogenesis of trophoblasts and trophoblast-endothelial interactions. Levels of angiogenic factors (VEGF and sFlt-1) in the supernatant were measured by ELISA, and the expression of cell cycle regulatory (cyclin D1) and invasive (MMP2, MMP3, MMP9) markers implicated in artery remodeling were measured by western blotting.
RESULTS: Ectopic expression of CCNG2 blocked the proliferation of HTR8/SVneo cells, as well as their abilities to form networks and integrate into human umbilical vein endothelial cells, whereas CCNG2 inhibition had the opposite effects. CCNG2 upregulation significantly reduced the expression of VEGF, cyclin D1, MMP2, MMP3, and MMP9, but enhanced the expression of sFlt-1. In contrast, CCNG2 downregulation had the opposite effects.
CONCLUSIONS: CCNG2 plays a critical role in trophoblast proliferation and trophoblast-endothelial cell interactions by significant affecting cell cycle, angiogenic, and invasive markers. CCNG2 may thus be a novel marker for the treatment of placental disorders.
Keywords: Cyclin G2, gestational trophoblastic disease, Cell Communication, Cell Line, Human Umbilical Vein Endothelial Cells, Trophoblasts
Background
Appropriate remodeling of spiral arteries is essential for successful fetal growth and pregnancy. During the first trimester of human pregnancy, extravillous trophoblasts (EVTs) differentiate into 2 invasive subpopulations: interstitial (inEVTs) and endovascular (enEVTs) EVTs. InEVTs invade the decidual stroma and the first third of the myometrium before finally reaching the outer aspects of the arterial walls. In contrast, enEVTs migrate across the inner wall of spiral arteries and adopt an endothelial-like phenotype, followed by trophoblast integration and the subsequent replacement of endothelial cells via their apoptosis and the catabolism of the extracellular matrix [1–6], a process called spiral artery remodeling. The vessels are characterized by high perfusion and low resistance to meet the requirements of the developing fetus [7].
Failure of spiral artery remodeling is closely associated with serious pregnancy-associated disorders, such as early pregnancy loss, fetal growth restriction, and preeclampsia. Preeclampsia is the major cause of maternal and perinatal mortality, with a worldwide incidence of 2–8% [8–13]. In addition, excessive invasion leads to placenta accrete [14]. However, the specific molecular mechanisms involved in this process have not been fully explored. Further research into possible regulators of trophoblast proliferation [15], differentiation [16], invasion [17], and angiogenesis [18] should enhance understanding of spiral artery remodeling, as well as placental development.
Cyclin G2 (CCNG2), which is encoded by the
Although evidence has suggested that CCNG2 may be involved in embryo implantation and trophoblast cell differentiation [31,32], the precise functions of CCNG2 in the remodeling of spiral arteries remain unclear. The present study was designed to examine the roles and potential mechanisms of CCNG2 in the regulation of trophoblast proliferation and trophoblast-endothelial cell interactions, and thereby identify a novel marker for the treatment of placenta-related diseases
Material and Methods
CELL CULTURE:
The human first trimester EVT cell line HTR8/SVneo was the kind gift of Dr. Charles Graham of Queen’s University, Kingston, Ontario, Canada [33]. Human umbilical vein endothelial cells (HUVECs) were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). HTR8/SVneo cells and HUVECs were cultured in Roswell Park Memorial Institute-1640 medium (RPMI-1640; Gibco, Carlsbad, CA, USA) and Dulbecco’s modified Eagle’s medium (DMEM; Gibco), respectively. Both media were supplemented with 100 IU/ml penicillin (Gibco), 100 mg/ml streptomycin (Gibco), and 10% (v/v) fetal bovine serum (FBS; Biological Industries, Kibbutz Beit Haemek, Israel), and both cell lines were cultured at 37°C with 5% CO2 in a humidified incubator.
LENTIVIRUS INFECTION:
To generate cell lines stably overexpressing CCNG2, HTR8/SVneo cells were infected with lentiviral particles carrying FLAG-tagged CCNG2 or control vector (GeneChem, Shanghai, China), yielding cells overexpressing CCNG2 (LV-CCNG2) and control cells (LV-NC), respectively. To generate CCNG2 knockdown cells using the CRISPR/Cas9 gene editing system, lentiviral Cas9, lentiviral sgRNA targeting the human
RNA EXTRACTION AND QRT-PCR:
Total RNA was extracted from infected HTR8/SVneo cells using TRIzol reagent according to the manufacturer’s protocol (Qiagen, CA, USA). cDNA was synthesized using a reverse transcription kit (RR036A; Takara, Tokyo, Japan). qRT-PCR was performed using a SYBR Green PCR kit (Takara) on a Roche LightCycler480 Real-Time PCR system and primers for
The level of expression of the CCNG2 gene was normalized to that of GAPDH, and relative expression of CCNG2 was calculated using the 2−ΔΔCt method [34].
WESTERN BLOT ANALYSIS:
Cells were harvested and lysed with radioimmunoprecipitation assay (RIPA) buffer supplemented with Protease Inhibitor Cocktail and PhosSTOP Phosphatase Inhibitor Cocktail (Roche, Basel, Switzerland). The protein concentrations of the cell lysate supernatants were quantified using a BCA Assay Kit (Beyotime, China). Equal amounts of protein samples were separated by SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). After being blocked, the membranes were incubated at 4°C overnight with one of the following primary antibodies: anti-CCNG2 (1: 1000; Abcam, USA), cyclin D1 (1: 1000; Abcam), anti-matrix metalloproteinase-2 (MMP2) (1: 1000; CST, USA), MMP3 (1: 1000; CST), anti-MMP9 (1: 1000; CST), or GAPDH (as loading control; 1: 5000; Proteintech, USA). After washing, the membranes were incubated with secondary horseradish peroxidase-conjugated anti-rabbit IgG antibody (1: 10000; Abcam) at room temperature for 1 h, with the bands subsequently visualized by chemiluminescence (DNR Bio-Imaging Systems, Jerusalem, Israel). Band intensity was quantified using ImageJ software (National Institutes of Health, USA) and was normalized to GAPDH expression.
CELL PROLIFERATION:
Cell proliferation was assessed using a Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, USA), according to the manufacturer’s instructions. Briefly, cells were seeded in 96-well plates at a density of 4×103 cells per well in 100 μl complete medium and incubated for 0, 1, 2, 3, 4, or 5 days. To each well was added 10 μl CCK-8 solution, followed by further incubation at 37°C for 3 h. The OD450 was measured by a microplate reader (Thermo Fisher Scientific, Waltham, MA, USA), and cell proliferation curves were plotted.
HTR8/SVNEO NETWORK FORMATION:
For Matrigel network formation assays, each well of a 96-well plate was coated with 40 μl ice-cold Matrigel (BD Biosciences, USA) and polymerized for 30 min at 37°C. HTR8/SVneo cells (4×104 cells/well) were seeded on the upper layer of the coated wells and cultured in 100 μl medium. Six hours later, the plates were imaged using a microscope (Nikon, Japan). Total network length and numbers of branch points were quantified using ImageJ software.
HUVEC LABELING:
HUVECs were labeled with red fluorescent CM-Dil (Yeasen, Shanghai, China), which stains viable cell membranes, according to the manufacturer’s protocol. Briefly, HUVECs with high confluency were collected, digested, washed, and dissociated into single cell suspensions at a concentration of 1 × 106 cells/ml. Prepared CM-Dil stock was added to each cell suspension at 2 μg/ml, followed by incubation at 37°C for 5 min and then at 4°C for another 15 min. Following centrifugation and removal of the supernatants, labeled cells were washed in PBS to eliminate residual fluorescent dye and cultured. Cell fluorescence was monitored under a fluorescence microscope (Olympus, Tokyo, Japan).
CO-CULTURE OF LABELED HUVECS AND HTR8/SVNEO CELLS ON MATRIGEL:
To assess the ability of HTR8/SVneo cells to integrate into endothelium, green fluorescent trophoblasts and red fluorescent endothelial cells were co-cultured as described, with minor modifications [35–38]. Briefly, 24-well culture plates were coated with undiluted Matrigel (BD Biosciences) (300 μl/well) and polymerized at 37°C for 30 min. Labeled HUVECs were seeded at 1.0×105 cells/well and grown for 4 h in DMEM to develop endothelial cellular networks. HTR8/SVneo cells (1.0×105 cells/well) were seeded onto the HUVEC monolayer and the 1: 1 mixtures were cultured in DMEM/RPIM-1640 medium for 24 h. Images were taken with a fluorescence microscope (Olympus). HTR8/SVneo cell integration into endothelial cells was calculated as the ratio of the green to the red fluorescent network formation area using ImageJ software.
ELISA:
Vascular endothelial growth factor (VEGF) and soluble fms-like tyrosine kinase receptor 1 (sFlt-1) concentrations in the conditioned media of infected HTR8/SVneo cells were determined with commercially available human VEGF and sFlt-1 ELISA kits (Uscn Life Science Inc., Wuhan, China), respectively, according to the manufacturer’s protocols. The optical density of each well at 450 nm was measured on a microplate reader (Thermo Fisher Scientific Inc., USA), with concentrations determined from standard curves. Results are shown as the concentration (pg/ml) compared with control.
STATISTICAL ANALYSIS:
Quantitative data are presented as the mean±standard deviation of triplicate determinations and compared using Student’s
Results
CONFIRMATION OF CCNG2 OVEREXPRESSION AND KNOCKDOWN:
To explore the effect of CCNG2 on the regulation of trophoblast cell function during the remodeling of the maternal spiral artery in vitro, CCNG2 was successfully overexpressed in HTR8/SVneo cells by infection with a lentivirus carrying the CCNG2 gene; similarly, CCNG2 expression was sufficiently silenced after treatment of these cells with Cas9/sgRNA vector. The overexpression and silencing of CCNG2 were verified at the mRNA and protein levels in whole cell lysates. The level of CCNG2 mRNA after infection of cells with the overexpression vector was 25.2-fold higher than control (P<0.001), whereas the level of CCNG2 mRNA after infection of cells with the knockdown vector was 9.1% that of control (P<0.05) (Figure 1). Similarly, CCNG2 protein levels in whole cell lysates following infection with the overexpression and knockdown vectors were 2.3-fold higher than (P<0.01) and 42.7% (P<0.01) of control, respectively.
CCNG2 BLOCKS THE PROLIFERATION OF HTR8/SVNEO CELLS:
CCK-8 assays of HTR8/SVneo cell proliferation following stable overexpression and silencing of CCNG2 showed that CCNG2 overexpression significantly reduced HTR8/SVneo cell proliferation, to 68.5% of control at 5 days (P<0.01), whereas CCNG2 knockdown enhanced cell proliferation to 1.3-fold higher than control at 5 days (P<0.05) (Figure 2). Taken together, these findings indicate that CCNG2 blocks the proliferation of trophoblast cells.
CCNG2 IMPAIRS THE FORMATION OF HTR8/SVNEO CELL NETWORKS:
Because the angiogenic property of trophoblast cells is critical for spiral artery remodeling, we also evaluated the effects of CCNG2 on the network formation capacity of HTR8/SVneo cells. Overexpression of CCNG2 significantly decreased the total network length to 23.3% of control (773±223 μm vs. 3318±361 μm, P<0.01) and reduced the number of branch points to 65.8% of control (12.7±2.1 vs. 19.3±1.2, P<0.05) (Figure 3). In contrast, knockdown of CCNG2 significantly increased the total network length to 1.4 times that of control (5173±400 μm vs. 3708±189 μm, P<0.05) and increased the number of branch points to 1.2 times that of control (22.3±1.5 vs. 18.7±0.6, P<0.05). Thus, these findings indicated that CCNG2 represses the angiogenic activity of trophoblast cells.
CCNG2 REDUCES THE CAPACITY OF HTR8/SVNEO CELLS TO INTEGRATE INTO HUVECS:
Green fluorescent-labeled trophoblast HTR8/SVneo cells and red fluorescent-labeled HUVECs were co-cultured on Matrigel to study the interaction of trophoblasts and endothelium. In the images, the capacity of HTR8/SVneo cells to integrate into HUVECs is represented by the ratio of green fluorescent to red fluorescent network areas. CCNG2 overexpression was found to reduce the capacity of HTR8/SVneo cells to replace endothelial networks to 27.6% of control (P<0.01), whereas CCNG2 knockdown increased the migration and integration of HTR8/SVneo cells into HUVECs by 2.3-fold relative to control (P<0.05) (Figure 4). These findings indicated that CCNG2 inhibits trophoblast integration into endothelial cells during spiral artery remodeling.
CCNG2 SUPPRESSES THE LEVEL OF VEGF BUT SIMULATES THAT OF SFLT-1:
To assess the impact of CCNG2 on trophoblast cell angiogenesis, we assessed its effects on the levels of the pro-angiogenic factor VEGF and the anti-angiogenic factor sFlt-1. Overexpression of CCNG2 reduced the concentration of VEGF in cell supernatants to 20.2% of control (8.5±2.9 pg/ml vs. 42.0±6.5 pg/ml, P<0.01), while increasing the concentration of sFlt-1 to 1.3-fold higher than control (362.4±8.4 pg/ml vs. 278.9±11.2 pg/ml, P<0.01) (Figure 5). Conversely, silencing of CCNG2 increased the concentration of VEGF to 1.7-fold higher than control (82.7±4.1 pg/ml vs. 49.4±6.7 pg/ml, P<0.05), while reducing the concentration of sFlt-1 to 65.5% of control (83.2±6.7 pg/ml vs. 127.1±7.5 pg/ml, P<0.05). Thus, CCNG2 can inhibit angiogenesis by suppressing VEGF expression and increasing sFlt-1 expression.
CCNG2 REDUCES THE EXPRESSION OF CYCLIN D1, MMP2, MMP3, AND MMP9:
To further confirm the potential molecular mechanism underlying the regulation of spiral artery remodeling by CCNG2, its effects on the levels of the cell cycle regulator cyclin D1 and the cell invasion markers MMP2, MMP3, and MMP9 were assessed by western blotting. CCNG2 overexpression significantly reduced the levels of expression of cyclin D1, MMP2, MMP3, and MMP9 proteins, whereas CCNG2 knockdown significantly increased the levels of all 4 (Figure 6). Taken together, these results indicate that CCNG2 participates in spiral artery remodeling by regulating cyclin D1 and MMPs.
Discussion
Successful fetal growth and pregnancy outcomes are determined by appropriate remodeling of spiral arteries during early pregnancy. The mechanism underlying the accurate remodeling of spiral arteries remains to be determined, although abnormally regulated trophoblasts likely play a crucial role in this process. The present study showed that CCNG2 could dramatically reduce the proliferative and angiogenic capacities of trophoblasts, as well as reducing their ability to integrate into endothelial cells. These findings suggest that CCNG2 is involved in the regulation of trophoblast cell differentiation to an endovascular trophoblast-like phenotype and the replacement of endothelial cells of uterine spiral arteries.
CCNG2 also has effects on the placenta. For example, a global analysis of gene expression profiles at the maternal-fetal interface found that CCNG2 expression in the placental basal plate region was 2.03-fold lower at term than during mid-pregnancy [39]. Similarly, transcriptome profiling of placental tissue identified a peak in placental
The present study showed that CCNG2 downregulated the expression in trophoblast cells of cyclin D1, a positive regulator of cell DNA synthesis and proliferation that promotes passage through the G1 to S checkpoint by associating with cyclin-dependent kinase (CDK) 4/6 [42]. Cyclin D1 is expressed in the nuclei of cytotrophoblasts in human placentas during the first trimester and is strongly expressed in the nuclei of endothelial cells during the third trimester [43], suggesting that cyclin D1 regulates the proliferation of trophoblast cells during early pregnancy and placental angiogenesis during late pregnancy. Moreover, placental cyclin D1 expression is decreased in women with fetal intrauterine growth restriction (IUGR) and preeclampsia complicated with IUGR [44], suggesting that abnormal regulation of cyclin D1 disturbs cell proliferation and compromises placental development. Because it is a link between growth signaling and cell division, cyclin D1 is a downstream target of several mitotic signals, such as AKT [45], ERK [46], NF-κB [47], and Wnt/β-catenin [48]. In addition to studying the relationship between CCNG2 and the Wnt/β-catenin signaling pathway [28,30], further research is required to determine whether other signaling pathways participate in the cyclin D1 abnormality mediated by CCNG2 during placental development.
During the physiological process of spiral artery remodeling, invasive EVTs produce extracellular matrix-degrading enzymes that proteolytically degrade musculoelastic layers around spiral arteries [49,50]. These enzymes are mainly induced by MMPs. In our study, the ability of CCNG2 to regulate the expression of MMP2, MMP3, and MMP9 was determined in HTR8/SVneo cells to assess the effects of cell invasion on CCNG2-mediated trophoblast-endothelial cell interactions. MMPs are a large family of zinc-dependent endopeptidases that are classified according to the proteins they degrade in the extracellular matrix. MMPs include the gelatinases MMP2 and MMP9 and the stromelysin MMP3 [51]. Downregulation of MMP2, MMP3, and MMP9 expression in preeclamptic placentas has been found to result in defective trophoblast invasion and impaired placentation [52–54]. Similarly, the present study showed that CCNG2 significantly reduced the levels of expression of MMP2, MMP3, and MMP9 in trophoblast cells, indicating that the ability of CCNG2 to interfere with the integration of trophoblasts into endothelial networks was due to its effects on invasive pathways, including the production of MMPs.
Our study revealed that overexpression of CCNG2 in HTR8/SVneo cells induced defects in network formation and integration into endothelial networks
Conclusions
These results demonstrate that CCNG2 participates in controlling trophoblast functions by impairing their proliferation, angiogenesis and integration into endothelial networks. This process is likely mediated by downstream effectors, including cell cycle proteins, MMPs, and the angiogenic factors VEGF and sFlt-1. CCNG2 may therefore be a regulator of trophoblast cell behavior that modulates placentation, suggesting a novel therapeutic approach to placental disorders.
Figures
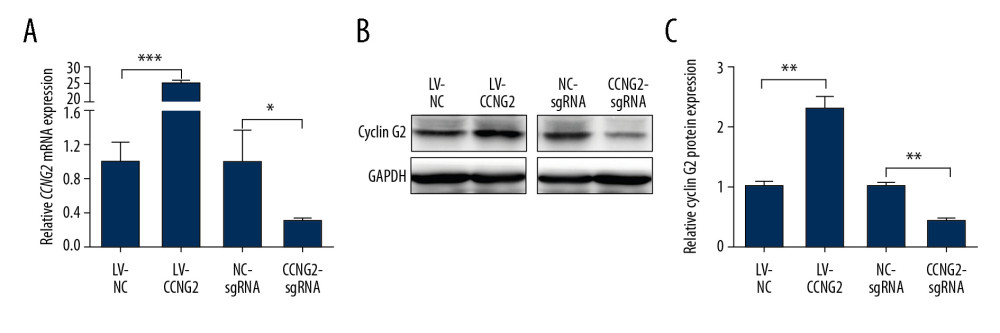 Figure 1. Effects of CCNG2 overexpression and knockdown in HTR8/SVneo cells. (A) qRT-PCR assays of relative CCNG2 mRNA expression showing its overexpression and knockdown in HTR8/SVneo cells. (B) Western blot assays of relative levels of CCNG2 protein showing its overexpression and knockdown in HTR8/SVneo cells. (C) Densitometric quantification of western blot results using ImageJ software. Each experiment was independently performed 3 times. * P<0.05, ** P<0.01, and *** P<0.001 vs. control.
Figure 1. Effects of CCNG2 overexpression and knockdown in HTR8/SVneo cells. (A) qRT-PCR assays of relative CCNG2 mRNA expression showing its overexpression and knockdown in HTR8/SVneo cells. (B) Western blot assays of relative levels of CCNG2 protein showing its overexpression and knockdown in HTR8/SVneo cells. (C) Densitometric quantification of western blot results using ImageJ software. Each experiment was independently performed 3 times. * P<0.05, ** P<0.01, and *** P<0.001 vs. control. 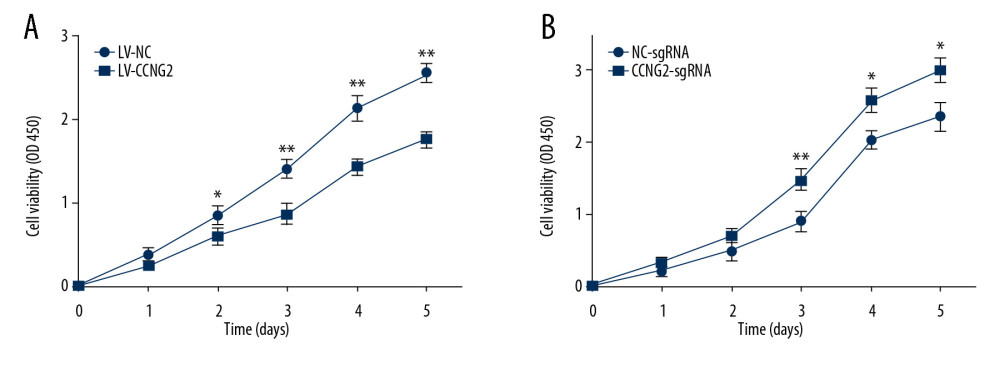 Figure 2. CCNG2 inhibits the proliferation of HTR8/SVneo cells. (A, B) CCK-8 assays showing the effects of CCNG2 overexpression (A) and knockdown (B) on the proliferation of HTR8/SVneo cells. Each experiment was independently performed 3 times. OD, optical density. * P<0.05 and ** P<0.01 vs. control.
Figure 2. CCNG2 inhibits the proliferation of HTR8/SVneo cells. (A, B) CCK-8 assays showing the effects of CCNG2 overexpression (A) and knockdown (B) on the proliferation of HTR8/SVneo cells. Each experiment was independently performed 3 times. OD, optical density. * P<0.05 and ** P<0.01 vs. control. 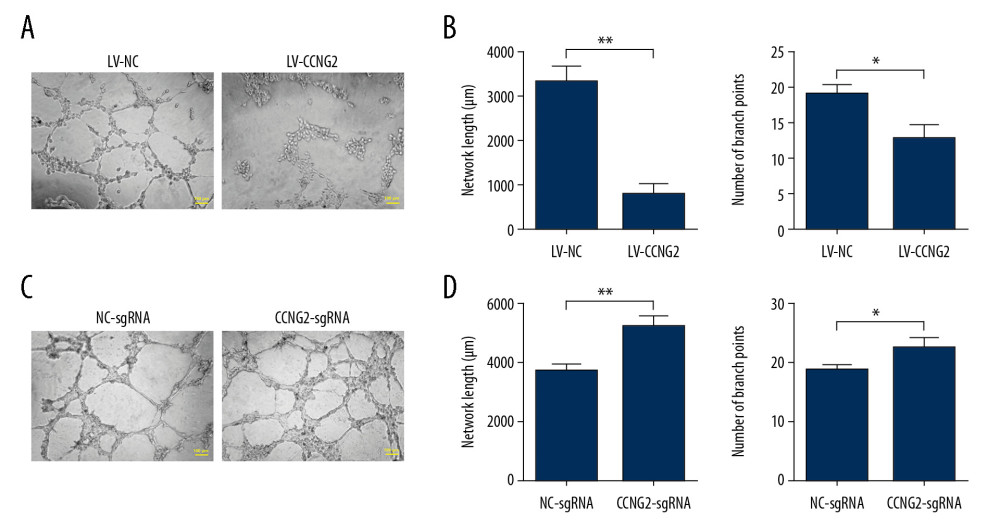 Figure 3. CCNG2 impairs the network formation of HTR8/SVneo cells. (A) Network formation assay to determine the angiogenic capacity of HTR8/SVneo cells overexpressing CCNG2. Original magnification×100. (B, C) Statistical analyses of total network length (B) and number of branch points (C) during network formation in cells overexpressing CCNG2, as determined using ImageJ software. (D) Network formation assay to determine the angiogenic capacity of HTR8/SVneo cells with CCNG2 silencing. Original magnification×100. (E, F) Statistical analyses of total network length (E) and number of branch points (F) during network formation in cells with CCNG2 knockdown, as determined using ImageJ software. Each experiment was independently performed 3 times. * P<0.05 and ** P<0.01 vs. control.
Figure 3. CCNG2 impairs the network formation of HTR8/SVneo cells. (A) Network formation assay to determine the angiogenic capacity of HTR8/SVneo cells overexpressing CCNG2. Original magnification×100. (B, C) Statistical analyses of total network length (B) and number of branch points (C) during network formation in cells overexpressing CCNG2, as determined using ImageJ software. (D) Network formation assay to determine the angiogenic capacity of HTR8/SVneo cells with CCNG2 silencing. Original magnification×100. (E, F) Statistical analyses of total network length (E) and number of branch points (F) during network formation in cells with CCNG2 knockdown, as determined using ImageJ software. Each experiment was independently performed 3 times. * P<0.05 and ** P<0.01 vs. control. 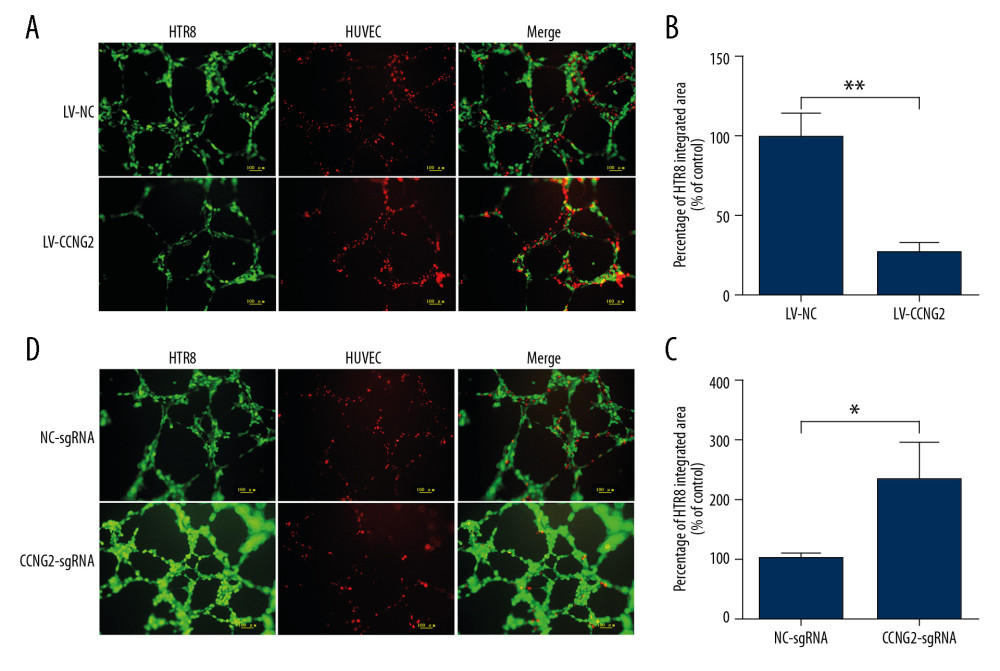 Figure 4. CCNG2 decreases the capacity of HTR8/SVneo cells to integrate into HUVECs. (A, C) Fluorescent images showing the 24-h co-culture of HUVECs with HTR8/SVneo cells overexpressing CCNG2 (A) and HTR8/SVneo cells with CCNG2 silencing (C). The left images show the cellular networks formed by green-stained HTR8/SVneo cells. The middle images show the cellular networks formed by red-stained HUVECs. The right images show the merged images. Original magnification×100. (B, D) Statistical analyses of the integration into HUVECs of HTR8/SVneo cells overexpressing CCNG2 (B) and HTR8/SVneo cells with CCNG2 silencing (D), as determined using ImageJ software. Each experiment was independently performed 3 times.* P<0.05 and ** P<0.01 vs. control.
Figure 4. CCNG2 decreases the capacity of HTR8/SVneo cells to integrate into HUVECs. (A, C) Fluorescent images showing the 24-h co-culture of HUVECs with HTR8/SVneo cells overexpressing CCNG2 (A) and HTR8/SVneo cells with CCNG2 silencing (C). The left images show the cellular networks formed by green-stained HTR8/SVneo cells. The middle images show the cellular networks formed by red-stained HUVECs. The right images show the merged images. Original magnification×100. (B, D) Statistical analyses of the integration into HUVECs of HTR8/SVneo cells overexpressing CCNG2 (B) and HTR8/SVneo cells with CCNG2 silencing (D), as determined using ImageJ software. Each experiment was independently performed 3 times.* P<0.05 and ** P<0.01 vs. control. 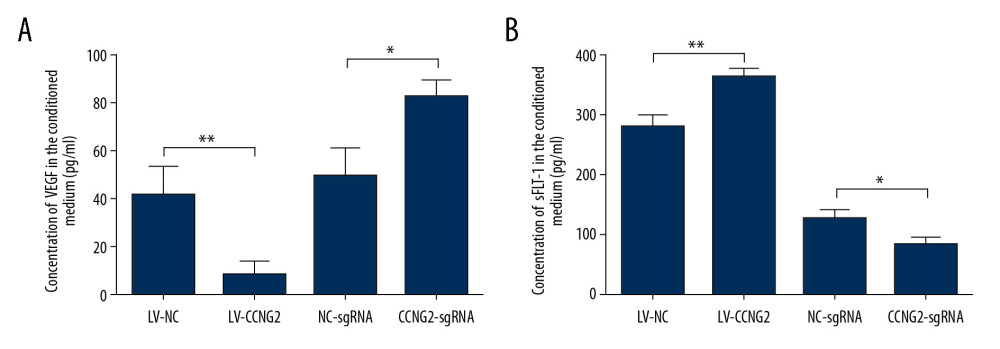 Figure 5. CCNG2 suppresses the expression of VEGF but increases that of sFlt-1. (A, B) Statistical analyses of the concentrations of VEGF (A) and sFlt-1 (B) in the conditioned media of infected HTR8/SVneo cells (pg/ml). Each experiment was independently performed 3 times. * P<0.05 and ** P<0.01 vs. control.
Figure 5. CCNG2 suppresses the expression of VEGF but increases that of sFlt-1. (A, B) Statistical analyses of the concentrations of VEGF (A) and sFlt-1 (B) in the conditioned media of infected HTR8/SVneo cells (pg/ml). Each experiment was independently performed 3 times. * P<0.05 and ** P<0.01 vs. control. 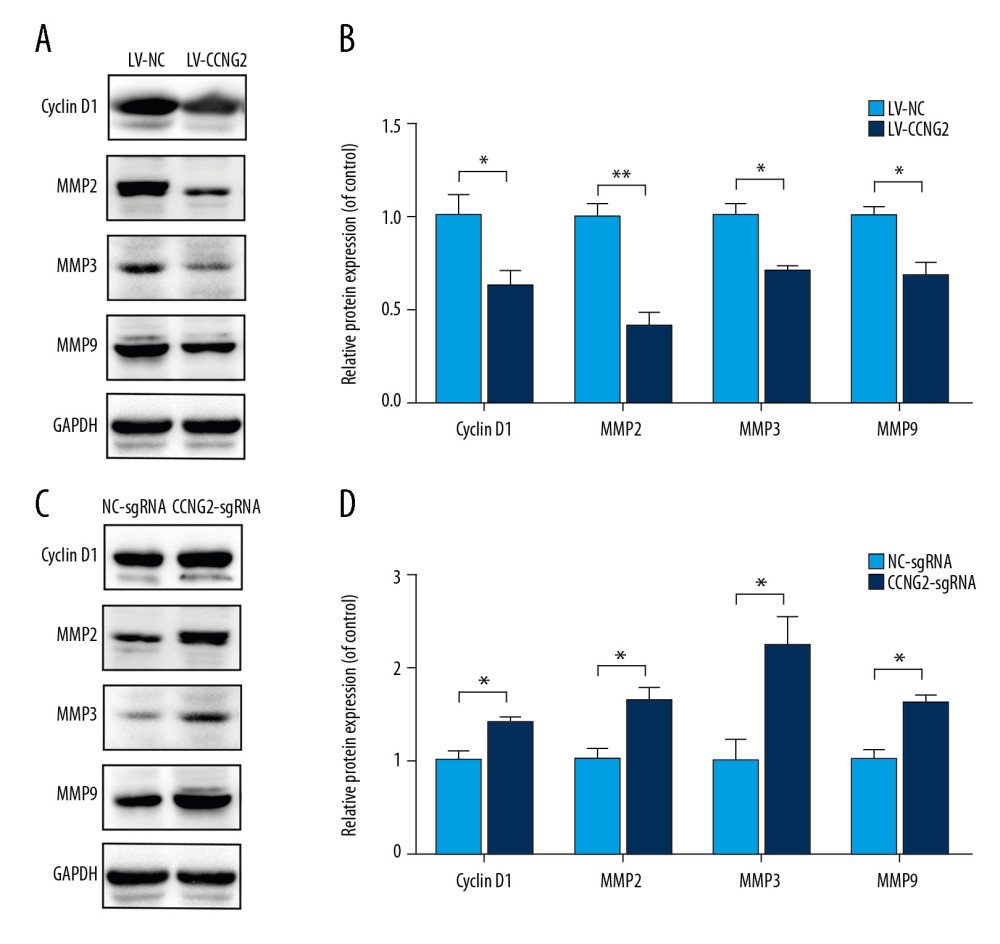 Figure 6. CCNG2 reduces the expression of cyclin D1, MMP2, MMP3, and MMP9. (A) Western blots showing the expression of cyclin D1, MMP2, MMP3, MMP9, and GAPDH in HTR8/SVneo cells overexpressing CCNG2. (B) Densitometric analysis of the results in A using ImageJ software. (C) Western blots showing the expression of cyclin D1, MMP2, MMP3, MMP9, and GAPDH in HTR8/SVneo cells with CCNG2 silencing. (D) Densitometric analysis of the results in C using ImageJ software. Each experiment was independently performed 3 times. * P<0.05 and ** P<0.01 vs. control.
Figure 6. CCNG2 reduces the expression of cyclin D1, MMP2, MMP3, and MMP9. (A) Western blots showing the expression of cyclin D1, MMP2, MMP3, MMP9, and GAPDH in HTR8/SVneo cells overexpressing CCNG2. (B) Densitometric analysis of the results in A using ImageJ software. (C) Western blots showing the expression of cyclin D1, MMP2, MMP3, MMP9, and GAPDH in HTR8/SVneo cells with CCNG2 silencing. (D) Densitometric analysis of the results in C using ImageJ software. Each experiment was independently performed 3 times. * P<0.05 and ** P<0.01 vs. control. References
1. Ashton SV, Whitley GS, Dash PR, Uterine spiral artery remodeling involves endothelial apoptosis induced by extravillous trophoblasts through Fas/FasL interactions: Arterioscler Thromb Vasc Biol, 2005; 25(1); 102-8
2. Harris LK, Smith SD, Keogh RJ, Trophoblast- and vascular smooth muscle cell-derived MMP-12 mediates elastolysis during uterine spiral artery remodeling: Am J Pathol, 2010; 177(4); 2103-15
3. Keogh RJ, Harris LK, Freeman A, Fetal-derived trophoblast use the apoptotic cytokine tumor necrosis factor-alpha-related apoptosis-inducing ligand to induce smooth muscle cell death: Circ Res, 2007; 100(6); 834-41
4. Whitley GS, Cartwright JE, Trophoblast-mediated spiral artery remodelling: A role for apoptosis: J Anat, 2009; 215(1); 21-26
5. Chakraborty C, Gleeson LM, McKinnon T, Lala PK, Regulation of human trophoblast migration and invasiveness: Can J Physiol Pharmacol, 2002; 80(2); 116-24
6. Zou Y, Jiang Z, Yu X, Upregulation of long noncoding RNA SPRY4-IT1 modulates proliferation, migration, apoptosis, and network formation in trophoblast cells HTR-8SV/neo: PLoS One, 2013; 8(11); e79598
7. Pijnenborg R, Vercruysse L, Hanssens M, The uterine spiral arteries in human pregnancy: Facts and controversies: Placenta, 2006; 27(9–10); 939-58
8. , ACOG Practice Bulletin No. 202 summary: Gestational hypertension and preeclampsia: Obstet Gynecol, 2019; 133(1); 211-14
9. Kaufmann P, Black S, Huppertz B, Endovascular trophoblast invasion: implications for the pathogenesis of intrauterine growth retardation and preeclampsia: Biol Reprod, 2003; 69(1); 1-7
10. Orozco AF, Jorgez CJ, Ramos-Perez WD, Placental release of distinct DNA-associated micro-particles into maternal circulation: Reflective of gestation time and preeclampsia: Placenta, 2009; 30(10); 891-97
11. van den Brule F, Berndt S, Simon N, Trophoblast invasion and placentation: Molecular mechanisms and regulation: Chem Immunol Allergy, 2005; 88; 163-80
12. Pijnenborg R, Vercruysse L, Hanssens M, Brosens I, Endovascular trophoblast and preeclampsia: A reassessment: Pregnancy Hypertens, 2011; 1(1); 66-71
13. Zhou Y, Damsky CH, Fisher SJ, Preeclampsia is associated with failure of human cytotrophoblasts to mimic a vascular adhesion phenotype. One cause of defective endovascular invasion in this syndrome?: J Clin Invest, 1997; 99(9); 2152-64
14. Duzyj CM, Buhimschi IA, Motawea H, The invasive phenotype of placenta accreta extravillous trophoblasts associates with loss of E-cadherin: Placenta, 2015; 36(6); 645-51
15. Ishihara N, Matsuo H, Murakoshi H, Increased apoptosis in the syncytiotrophoblast in human term placentas complicated by either preeclampsia or intrauterine growth retardation: Am J Obstet Gynecol, 2002; 186(1); 158-66
16. Zhang Y, Zou Y, Wang W, Down-regulated long non-coding RNA MEG3 and its effect on promoting apoptosis and suppressing migration of trophoblast cells: J Cell Biochem, 2015; 116(4); 542-50
17. Waddell JM, Evans J, Jabbour HN, Denison FC, CTGF expression is up-regulated by PROK1 in early pregnancy and influences HTR-8/Svneo cell adhesion and network formation: Hum Reprod, 2011; 26(1); 67-75
18. Maynard SE, Min JY, Merchan J, Excess placental soluble fms-like tyrosine kinase 1 (sFlt1) may contribute to endothelial dysfunction, hypertension, and proteinuria in preeclampsia: J Clin Invest, 2003; 111(5); 649-58
19. Horne MC, Donaldson KL, Goolsby GL, Cyclin G2 is up-regulated during growth inhibition and B cell antigen receptor-mediated cell cycle arrest: J Biol Chem, 1997; 272(19); 12650-61
20. Zimmermann M, Arachchige-Don AS, Donaldson MS, Elevated cyclin G2 expression intersects with DNA damage checkpoint signaling and is required for a potent G2/M checkpoint arrest response to doxorubicin: J Biol Chem, 2012; 287(27); 22838-53
21. Adorno M, Cordenonsi M, Montagner M, A mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis: Cell, 2009; 137(1); 87-98
22. Ito Y, Yoshida H, Uruno T, Decreased expression of cyclin G2 is significantly linked to the malignant transformation of papillary carcinoma of the thyroid: Anticancer Res, 2003; 23(38); 2335-38
23. Kim Y, Shintani S, Kohno Y, Cyclin G2 dysregulation in human oral cancer: Cancer Res, 2004; 64(24); 8980-86
24. Hasegawa S, Nagano H, Konno M, Cyclin G2: A novel independent prognostic marker in pancreatic cancer: Oncol Lett, 2015; 10(5); 2986-90
25. Wang S, Zeng Y, Zhou JM, MicroRNA-1246 promotes growth and metastasis of colorectal cancer cells involving CCNG2 reduction: Mol Med Rep, 2016; 13(1); 273-80
26. Choi MG, Noh JH, An JY, Expression levels of cyclin G2, but not cyclin E, correlate with gastric cancer progression: J Surg Res, 2009; 157(2); 168-74
27. Jia JS, Xu SR, Ma JExpression of cyclin G2 mRNA in patients with acute leukemia and its clinical significance]: Zhongguo Shi Yan Xue Ye Xue Za Zhi, 2005; 13(2); 254-59 [in Chinese]
28. Gao J, Zhao C, Liu Q, Cyclin G2 suppresses Wnt/β-catenin signaling and inhibits gastric cancer cell growth and migration through Dapper1: J Exp Clin Cancer Res, 2018; 37(1); 317
29. Li S, Gao J, Zhuang X, Cyclin G2 inhibits the Warburg effect and tumour progression by suppressing LDHA phosphorylation in glioma: Int J Biol Sci, 2019; 15(3); 544-55
30. Zhao C, Gao J, Li S, Cyclin G2 regulates canonical Wnt signalling via interaction with Dapper1 to attenuate tubulointerstitial fibrosis in diabetic nephropathy: J Cell Mol Med, 2020; 24(5); 2749-60
31. Yue L, Daikoku T, Hou X, Cyclin G1 and cyclin G2 are expressed in the periimplantation mouse uterus in a cell-specific and progesterone-dependent manner: Evidence for aberrant regulation with Hoxa-10 deficiency: Endocrinology, 2005; 146(5); 2424-33
32. Nadeem U, Ye G, Salem M, Peng C, MicroRNA-378a-5p targets cyclin G2 to inhibit fusion and differentiation in BeWo cells: Biol Reprod, 2014; 91(3); 76
33. Graham CH, Hawley TS, Hawley RG, Establishment and characterization of first trimester human trophoblast cells with extended lifespan: Exp Cell Res, 1993; 206(2); 204-11
34. Livak KJ, Schmittgen TD, Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method: Methods, 2001; 25(4); 402-8
35. Xu B, Charlton F, Makris A, Hennessy A, Nitric oxide (NO) reversed TNF-α inhibition of trophoblast interaction with endothelial cellular networks: Placenta, 2014; 35(6); 417-21
36. Xu B, Shanmugalingam R, Chau K: J Reprod Immunol, 2017; 124; 54-61
37. Xu B, Charlton F, Makris A, Hennessy A: J Hypertens, 2014; 32(5); 1075-83 discussion 1083
38. Xu B, Bobek G, Makris A, Hennessy A, Antihypertensive methyldopa, labetalol, hydralazine, and clonidine reversed tumour necrosis factor-α inhibited endothelial nitric oxide synthase expression in endothelial-trophoblast cellular networks: Clin Exp Pharmacol Physiol, 2017; 44(3); 421-27
39. Winn VD, Haimov-Kochman R, Paquet AC, Gene expression profiling of the human maternal-fetal interface reveals dramatic changes between midgestation and term: Endocrinology, 2007; 148(3); 1059-79
40. Uuskula L, Mannik J, Rull K, Mid-gestational gene expression profile in placenta and link to pregnancy complications: PLoS One, 2012; 7(11); e49248
41. Gates KC, Goetzmann LN, Cantlon JD, Effect of proline rich 15-deficiency on trophoblast viability and survival: PLoS One, 2017; 12(4); e0174976
42. Strauss M, Lukas J, Bartek J, Unrestricted cell cycling and cancer: Nat Med, 1995; 1(12); 1245-46
43. De Falco M, Fedele V, Cobellis L, Pattern of expression of cyclin D1/CDK4 complex in human placenta during gestation: Cell Tissue Res, 2004; 317(2); 187-94
44. Yung HW, Calabrese S, Hynx D, Evidence of placental translation inhibition and endoplasmic reticulum stress in the etiology of human intrauterine growth restriction: Am J Pathol, 2008; 173(2); 451-62
45. Hinz N, Jucker M, Distinct functions of AKT isoforms in breast cancer: A comprehensive review: Cell Commun Signal, 2019; 17(1); 154
46. Luo X, Liu J, Zhou H, Chen L, Apelin/APJ system: A critical regulator of vascular smooth muscle cell: J Cell Physiol, 2018; 233(7); 5180-88
47. Tilborghs S, Corthouts J, Verhoeven Y, The role of nuclear ractor-kappa B signaling in human cervical cancer: Crit Rev Oncol Hematol, 2017; 120; 141-50
48. Niehrs C, Acebron SP, Mitotic and mitogenic Wnt signalling: EMBO J, 2012; 31(12); 2705-13
49. Hazan AD, Smith SD, Jones RL: Am J Pathol, 2010; 177(2); 1017-30
50. Wallace AE, Fraser R, Cartwright JE, Extravillous trophoblast and decidual natural killer cells: A remodelling partnership: Hum Reprod Update, 2012; 18(4); 458-71
51. Zhu JY, Pang ZJ, Yu YH, Regulation of trophoblast invasion: The role of matrix metalloproteinases: Rev Obstet Gynecol, 2012; 5(3–4); e137-43
52. Reister F, Kingdom JCP, Ruck P, Altered protease expression by periarterial trophoblast cells in severe early-onset preeclampsia with IUGR: J Perinat Med, 2006; 34(4); 272-79
53. Zhu JY, Zhong M, Pang ZJ, Yu YH, Dysregulated expression of matrix metalloproteinases and their inhibitors may participate in the pathogenesis of pre-eclampsia and fetal growth restriction: Early Hum Dev, 2014; 90(10); 657-64
54. Espino Y, Sosa S, Flores-Pliego A, Espejel-Nunez A, New insights into the role of matrix metalloproteinases in preeclampsia: Int J Mol Sci, 2017; 18(7); 1448
55. Leanos-Miranda A, Campos-Galicia I, Berumen-Lechuga MG, Circulating angiogenic factors and the risk of preeclampsia in systemic lupus erythematosus pregnancies: J Rheumatol, 2015; 42(7); 1141-49
56. Farina A, Sekizawa A, De Sanctis P, Gene expression in chorionic villous samples at 11 weeks’ gestation from women destined to develop preeclampsia: Prenat Diagn, 2008; 28(10); 956-61
57. He Y, Smith SK, Day KA, Alternative splicing of vascular endothelial growth factor (VEGF)-R1 (FLT-1) pre-mRNA is important for the regulation of VEGF activity: Mol Endocrinol, 1999; 13(4); 537-45
58. Ferrara N, Gerber HP, LeCouter J, The biology of VEGF and its receptors: Nat Med, 2003; 9(6); 669-76
Figures
 Figure 1. Effects of CCNG2 overexpression and knockdown in HTR8/SVneo cells. (A) qRT-PCR assays of relative CCNG2 mRNA expression showing its overexpression and knockdown in HTR8/SVneo cells. (B) Western blot assays of relative levels of CCNG2 protein showing its overexpression and knockdown in HTR8/SVneo cells. (C) Densitometric quantification of western blot results using ImageJ software. Each experiment was independently performed 3 times. * P<0.05, ** P<0.01, and *** P<0.001 vs. control.
Figure 1. Effects of CCNG2 overexpression and knockdown in HTR8/SVneo cells. (A) qRT-PCR assays of relative CCNG2 mRNA expression showing its overexpression and knockdown in HTR8/SVneo cells. (B) Western blot assays of relative levels of CCNG2 protein showing its overexpression and knockdown in HTR8/SVneo cells. (C) Densitometric quantification of western blot results using ImageJ software. Each experiment was independently performed 3 times. * P<0.05, ** P<0.01, and *** P<0.001 vs. control. Figure 2. CCNG2 inhibits the proliferation of HTR8/SVneo cells. (A, B) CCK-8 assays showing the effects of CCNG2 overexpression (A) and knockdown (B) on the proliferation of HTR8/SVneo cells. Each experiment was independently performed 3 times. OD, optical density. * P<0.05 and ** P<0.01 vs. control.
Figure 2. CCNG2 inhibits the proliferation of HTR8/SVneo cells. (A, B) CCK-8 assays showing the effects of CCNG2 overexpression (A) and knockdown (B) on the proliferation of HTR8/SVneo cells. Each experiment was independently performed 3 times. OD, optical density. * P<0.05 and ** P<0.01 vs. control. Figure 3. CCNG2 impairs the network formation of HTR8/SVneo cells. (A) Network formation assay to determine the angiogenic capacity of HTR8/SVneo cells overexpressing CCNG2. Original magnification×100. (B, C) Statistical analyses of total network length (B) and number of branch points (C) during network formation in cells overexpressing CCNG2, as determined using ImageJ software. (D) Network formation assay to determine the angiogenic capacity of HTR8/SVneo cells with CCNG2 silencing. Original magnification×100. (E, F) Statistical analyses of total network length (E) and number of branch points (F) during network formation in cells with CCNG2 knockdown, as determined using ImageJ software. Each experiment was independently performed 3 times. * P<0.05 and ** P<0.01 vs. control.
Figure 3. CCNG2 impairs the network formation of HTR8/SVneo cells. (A) Network formation assay to determine the angiogenic capacity of HTR8/SVneo cells overexpressing CCNG2. Original magnification×100. (B, C) Statistical analyses of total network length (B) and number of branch points (C) during network formation in cells overexpressing CCNG2, as determined using ImageJ software. (D) Network formation assay to determine the angiogenic capacity of HTR8/SVneo cells with CCNG2 silencing. Original magnification×100. (E, F) Statistical analyses of total network length (E) and number of branch points (F) during network formation in cells with CCNG2 knockdown, as determined using ImageJ software. Each experiment was independently performed 3 times. * P<0.05 and ** P<0.01 vs. control. Figure 4. CCNG2 decreases the capacity of HTR8/SVneo cells to integrate into HUVECs. (A, C) Fluorescent images showing the 24-h co-culture of HUVECs with HTR8/SVneo cells overexpressing CCNG2 (A) and HTR8/SVneo cells with CCNG2 silencing (C). The left images show the cellular networks formed by green-stained HTR8/SVneo cells. The middle images show the cellular networks formed by red-stained HUVECs. The right images show the merged images. Original magnification×100. (B, D) Statistical analyses of the integration into HUVECs of HTR8/SVneo cells overexpressing CCNG2 (B) and HTR8/SVneo cells with CCNG2 silencing (D), as determined using ImageJ software. Each experiment was independently performed 3 times.* P<0.05 and ** P<0.01 vs. control.
Figure 4. CCNG2 decreases the capacity of HTR8/SVneo cells to integrate into HUVECs. (A, C) Fluorescent images showing the 24-h co-culture of HUVECs with HTR8/SVneo cells overexpressing CCNG2 (A) and HTR8/SVneo cells with CCNG2 silencing (C). The left images show the cellular networks formed by green-stained HTR8/SVneo cells. The middle images show the cellular networks formed by red-stained HUVECs. The right images show the merged images. Original magnification×100. (B, D) Statistical analyses of the integration into HUVECs of HTR8/SVneo cells overexpressing CCNG2 (B) and HTR8/SVneo cells with CCNG2 silencing (D), as determined using ImageJ software. Each experiment was independently performed 3 times.* P<0.05 and ** P<0.01 vs. control. Figure 5. CCNG2 suppresses the expression of VEGF but increases that of sFlt-1. (A, B) Statistical analyses of the concentrations of VEGF (A) and sFlt-1 (B) in the conditioned media of infected HTR8/SVneo cells (pg/ml). Each experiment was independently performed 3 times. * P<0.05 and ** P<0.01 vs. control.
Figure 5. CCNG2 suppresses the expression of VEGF but increases that of sFlt-1. (A, B) Statistical analyses of the concentrations of VEGF (A) and sFlt-1 (B) in the conditioned media of infected HTR8/SVneo cells (pg/ml). Each experiment was independently performed 3 times. * P<0.05 and ** P<0.01 vs. control. Figure 6. CCNG2 reduces the expression of cyclin D1, MMP2, MMP3, and MMP9. (A) Western blots showing the expression of cyclin D1, MMP2, MMP3, MMP9, and GAPDH in HTR8/SVneo cells overexpressing CCNG2. (B) Densitometric analysis of the results in A using ImageJ software. (C) Western blots showing the expression of cyclin D1, MMP2, MMP3, MMP9, and GAPDH in HTR8/SVneo cells with CCNG2 silencing. (D) Densitometric analysis of the results in C using ImageJ software. Each experiment was independently performed 3 times. * P<0.05 and ** P<0.01 vs. control.
Figure 6. CCNG2 reduces the expression of cyclin D1, MMP2, MMP3, and MMP9. (A) Western blots showing the expression of cyclin D1, MMP2, MMP3, MMP9, and GAPDH in HTR8/SVneo cells overexpressing CCNG2. (B) Densitometric analysis of the results in A using ImageJ software. (C) Western blots showing the expression of cyclin D1, MMP2, MMP3, MMP9, and GAPDH in HTR8/SVneo cells with CCNG2 silencing. (D) Densitometric analysis of the results in C using ImageJ software. Each experiment was independently performed 3 times. * P<0.05 and ** P<0.01 vs. control. In Press
05 Mar 2024 : Clinical Research
Muscular Function Recovery from General Anesthesia in 132 Patients Undergoing Surgery with Acceleromyograph...Med Sci Monit In Press; DOI: 10.12659/MSM.942780
05 Mar 2024 : Clinical Research
Effects of Thermal Insulation on Recovery and Comfort of Patients Undergoing Holmium Laser LithotripsyMed Sci Monit In Press; DOI: 10.12659/MSM.942836
05 Mar 2024 : Clinical Research
Role of Critical Shoulder Angle in Degenerative Type Rotator Cuff Tears: A Turkish Cohort StudyMed Sci Monit In Press; DOI: 10.12659/MSM.943703
06 Mar 2024 : Clinical Research
Comparison of Outcomes between Single-Level and Double-Level Corpectomy in Thoracolumbar Reconstruction: A ...Med Sci Monit In Press; DOI: 10.12659/MSM.943797
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952








