08 October 2020: Database Analysis
ATP6V1G3 Acts as a Key Gene in Recurrent Spontaneous Abortion: An Integrated Bioinformatics Analysis
Yihong Chen1BCE*, Jifen Hu1AEGDOI: 10.12659/MSM.927537
Med Sci Monit 2020; 26:e927537
Abstract
BACKGROUND: The molecular mechanism of recurrent spontaneous abortion is unclear. It has been suggested that dysregulated genes participate in the pathogenesis of recurrent spontaneous abortion. The aim of this study was to identify the differentially expressed genes (DEGs) and pathways in recurrent spontaneous abortion.
MATERIAL AND METHODS: Gene expression data series of GSE22490 and GSE26787 were obtained from the GEO database to identify the differentially expressed genes between patients with recurrent miscarriage (Case group) and patients with uncomplicated pregnancies matched for gestational age (Control group). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEEG) were applied to enrich the biological functions and pathways of the identified differentially expressed genes. A protein–protein interaction (PPI) network was constructed thorough the STRING database. Thirty-one cases of recurrent spontaneous abortion (Case group) and 30 cases of artificial abortion (Control group) were included in the study. The protein expression of hub genes in the villi and decidua tissue of the 2 groups was detected by immunohistochemical assay.
RESULTS: Forty-six DEGs were identified with the enriched biological function mainly in the aspects of glutamate secretion and positive regulation of synapse assembly. KEGG pathway analysis indicated the dysregulated genes were only enriched in the glutamatergic synapse pathway. In the PPI network, 83 nodes and 273 edges with the average node degreed of 6.58 were enriched. The hub gene (ATP6V1G3) of the included 46 genes was identified using Cytohubba software. In the Case group, the high expression of ATP6V1G3 protein was detected in 13 (43.3%) and 10 (33.3%) for placental villus and decidual tissue, respectively. However, the high expression rate in the Control group was 23.3% and 16.7% for placental villus and decidual tissue, respectively. The ATP6V1G3 protein high expression rate was not significantly different between the Case and Control groups (P>0.05).
CONCLUSIONS: We found differential gene expression profiles in villous and decidual tissues between patients with recurrent miscarriage vs. those with uncomplicated pregnancies. Upregulation of the ATP6V1G3 gene may play an important role in the development of recurrent miscarriage.
Keywords: Abortion, Habitual, Databases, Bibliographic, Computational Biology, Databases, Nucleic Acid, Gene Expression Regulation, Enzymologic, Pregnancy, Protein Interaction Maps, Vacuolar Proton-Translocating ATPases
Background
Recurrent spontaneous abortion is defined as 3 or more consecutive pregnancy losses at less than 12 weeks gestation [1]. Some factors related with this condition have been identified, such as chromosome abnormality, malformation of reproductive organs, hormonal imbalance, and infection and/or inflammation [2–5]. Immune rejection of maternal and fetal boundaries related to imbalance of immune tolerance has also been suggested to be a risk factor for recurrent spontaneous abortion [6]. Immune rejection was also reported to be a cause, but 50–75% of recurrent miscarriage are unexplained [7]. Furthermore, the molecular mechanism of recurrent spontaneous abortion is yet to be determined. Publications indicated that most recurrent spontaneous abortions were mainly or partially due to genetic factors [8–10]. In recent years, numerous microarrays and sequencing data were collated and stored in databases such as GEO, TGCA, Kaplan Meier Plotter, and STING databases. Clinical features and relevant data in these databases can be freely downloaded and further analyzed [11,12] by relevant statistical software or online analysis tools. The open assess data proved a useful platform for investigation of recurrent spontaneous abortion. The present study assessed the dysregulated genes associated with development of recurrent spontaneous abortion and further investigated their biological function.
Material and Methods
DATA SERIES RELEVANT TO RECURRENT ABORTION:
The data series relevant to recurrent spontaneous abortion were screened in the GEO database (https://www.ncbi.nlm.nih.gov/geo/). Two data series, GSE22490 [13] and GSE26787 [14], relevant to differentially expressed genes between recurrent miscarriage (Case group) and uncomplicated pregnancies matched for gestational age (Control group) were identified. The biological function analysis of the differentially expressed genes was performed using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEEG). The protein–protein interaction (PPI) network of the differentially expressed genes was constructed using the STRING database. Hub genes were identified using Cytohubba software.
DYSREGULATED GENES IDENTIFICATION AND FUNCTION ENRICHMENT:
The dysregulated genes in each data series (GSE22490 and GSE26787) were first screened by using the fold change (recurrent miscarriage
PPI NETWORK ANALYSIS:
The PPI network of the 46 dysregulated genes was constructed using the STRING database through input the 46 genes in Homo sapiens.
IMMUNOHISTOCHEMISTRY ASSAY:
Thirty-one cases of recurrent spontaneous abortion were collected from the Gynecology Department of the First Affiliated Hospital of Fujian Medical University as the recurrent spontaneous abortion (Case) group. We also collected data from 30 patients who decided to terminate their pregnancy because of unintended pregnancy and who underwent induced abortion. The Control group was limited to patients who were more than 18 years old and did not had a recurrent spontaneous abortion history and had no family history of genetic diseases. The protein expression of ATP6V1G3 in the villi and decidua tissue of the 2 groups was detected by immunohistochemical assay. The paraffin specimens were cut into slices 4–7 μm in thickness and soaked in xylene for 10 min, absolute ethanol for 5 min, and 95%, 80%, and 79% ethanol for 5 min. After dewaxing, specimens were incubated in 3% H2O2 in the dark for 15 min to inactivate endogenous peroxidase, after which 0.01% mol/L sodium citrate was added at 95°C for 15 min for antigen repair. We used 100 μl normal serum containing 10% for antigen sealing. The primary antibody was diluted with 10% serum, and then dropped on the tissue at 4°C and incubated overnight. After washing with PBS and DAB staining, the slice was sealed and prepared for microscopic examination. This study was approved by Ethics Committee of the First Affiliated Hospital of Fujian Medical University. Signed informed consent was obtained from all included subjects.
Results
IDENTIFICATION OF DYSREGULATED GENES:
Forty-six dysregulated genes were identified between the Case group and the Control group (Figure 1), as shown in the heat map in Figure 2.
GO AND KEGG ANALYSIS:
Gene ontology analysis indicated that the 46 DEGs were enriched in glutamate secretion, positive regulation of synapse assembly, T cell receptor signaling pathway, ion transmembrane transport, chemical synaptic transmission, and immune response in the aspect of biological process (BP). For the molecular function (MF), the dysregulated genes were enriched in MHC class II receptor activity, high-affinity glutamate transmembrane transporter activity, L-glutamate transmembrane transporter activity, water channel activity, amino acid transmembrane transporter activity, symporter activity, and transporter activity. KEGG pathway analysis indicated the dysregulated genes were only enriched in the glutamatergic synapse pathway (Figure 3).
PPI NETWORK AND HUB GENE ANALYSIS:
Protein–protein interaction (PPI) network was analyzed by STRING database, and 83 nodes and 273 edges with the average node degreed of 6.58 were established (Figure 4). Five hub genes (ATP6V1G3, ATP6V1E1, ATP6VOD2, ATP6VOB, and ATP6V1B1) of the 46included genes were identified using Cytohubba (Figure 5).
ATP6V1G3 PROTEIN EXPRESSION:
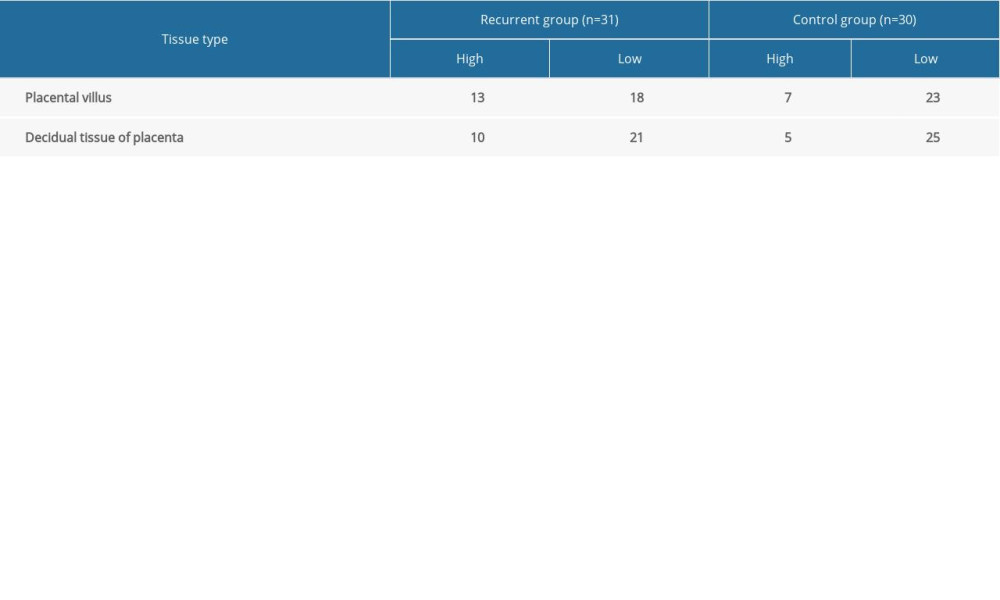
ATP6V1G3 protein was mainly expressed in the cytoplasm with brownish stain (Figure 6). In Case group, high expression of ATP6V1G3 protein was detected in 13 (43.3%) and 10 (33.3%) for placental villus and decidual tissue, respectively. However, the high expression rate in the Control group was 23.3% and 16.7% for placental villus and decidual tissue, respectively. The ATP6V1G3 protein high expression rates were not significantly different between the Case and Control groups (P>0.05) (Table 1).
Discussion
Recurrent spontaneous abortion (RSA) is a common complication of pregnancy, with an incidence rate of 1–5% [15]. Due to difficulties in treatment and high medical costs, it imposes a heavy family burden for couples who are eager to have children. Its etiology is complex and the known causes are mainly genetic factors, anatomical abnormalities of the reproductive tract, endocrine abnormalities, immune dysfunction, infectious factors, and thrombotic factors and external environmental factors [16,17]. However, about half of the recurrent spontaneous abortions did not have clear causes. Recurrent spontaneous abortion is generally divided into 2 types: non-immune recurrent spontaneous abortion and immune recurrent spontaneous abortion. The etiology of non-immune recurrent spontaneous abortion has been identified, including anatomical factors such as genital tract malformation (5%), endocrine factors (20%), infectious factors (5%), and genetic and chromosomal factors (5%). Immune recurrent spontaneous abortion mainly includes the autoimmune type, which is generally caused by anti-phospholipid antibody (APA), and it is part of the anti-phospholipid antibody syndrome (APS). However, the molecular mechanism of recurrence spontaneous abortion was not completely clear. It is unknown how many genes are involved in the recurrence spontaneous abortion, and their pathway is not known [18,19].
In this present study, we selected 2 gene chip datasets on differential expression between placental tissue of recurrent spontaneous abortion patients and placental tissue of induced abortion in the GEO database. We identified found 46 overlapped genes of the 2 datasets with differential expression. These 46 genes are mainly enriched in the biological function of glutamate secretion, positive regulation of synapse assembly, T cell receptor signaling pathway, MHC class II receptor activity, high-affinity glutamate trans membrane transporter activity, L-glutamate transmembrane transporter activity, and transporter activity. KEGG pathway analysis indicated the 46 dysregulated genes were only enriched in the glutamatergic synapse pathway. This gene panel of 46 differentially expressed genes may play a key role in the development of recurrence of recurrent spontaneous abortion and could be used as a biomarker for prediction of recurrent spontaneous abortion. In addition, we also found that in the Case group, the high expression of ATP6V1G3 protein was detected in 13 (43.3%) and 10 (33.3%) for placental villus and decidual tissue, respectively. The high expression rate in the Control group was 23.3% and 16.7% for placental villus and decidual tissue, respectively. The ATP6V1G3 protein high expression rate was not significantly different between the Case and the Control groups (
Conclusions
We found differential gene expression profiles between the Case group and th Control group, which may provide useful information for further research on differential gene expression in recurrent spontaneous abortion. Upregulation of the gene
Figures
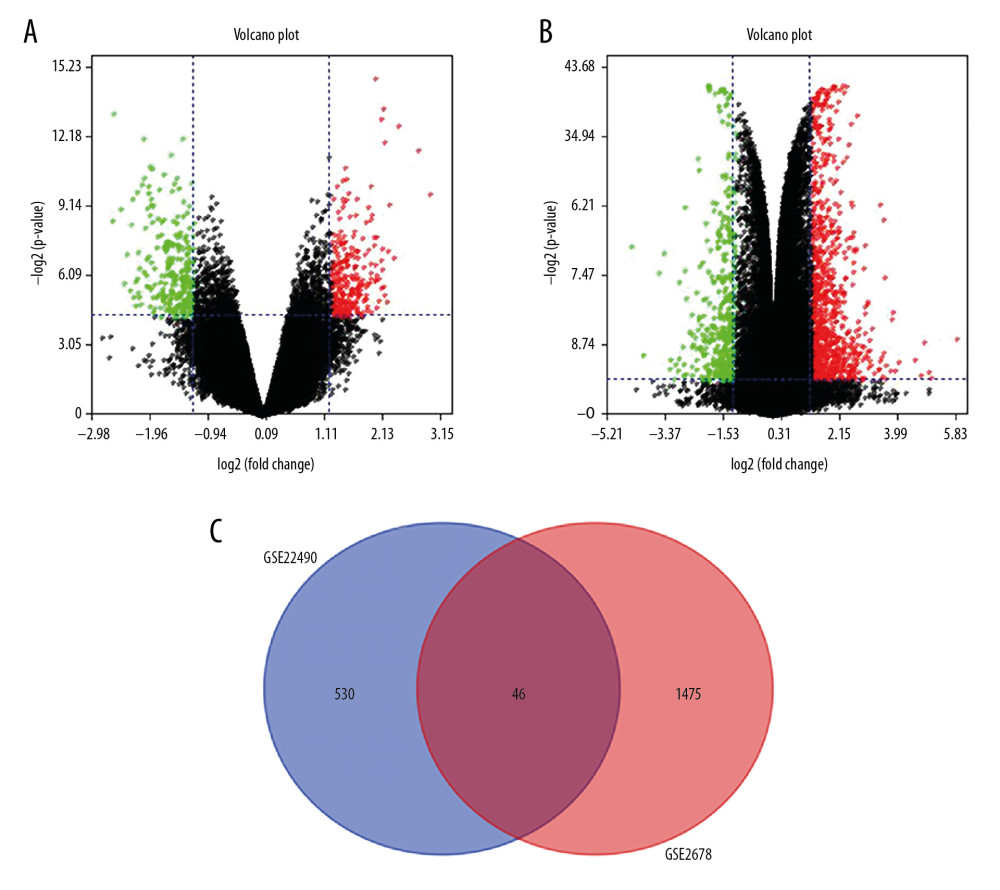 Figure 1. Identification of differentially expressed genes. We identified 46 overlapping dysregulated genes through GSE22490 and GSE2678 (A: Volcano plot of GSE22490; B: Volcano plot of GSE26787; C: Venn plot of the overlapping genes).
Figure 1. Identification of differentially expressed genes. We identified 46 overlapping dysregulated genes through GSE22490 and GSE2678 (A: Volcano plot of GSE22490; B: Volcano plot of GSE26787; C: Venn plot of the overlapping genes). 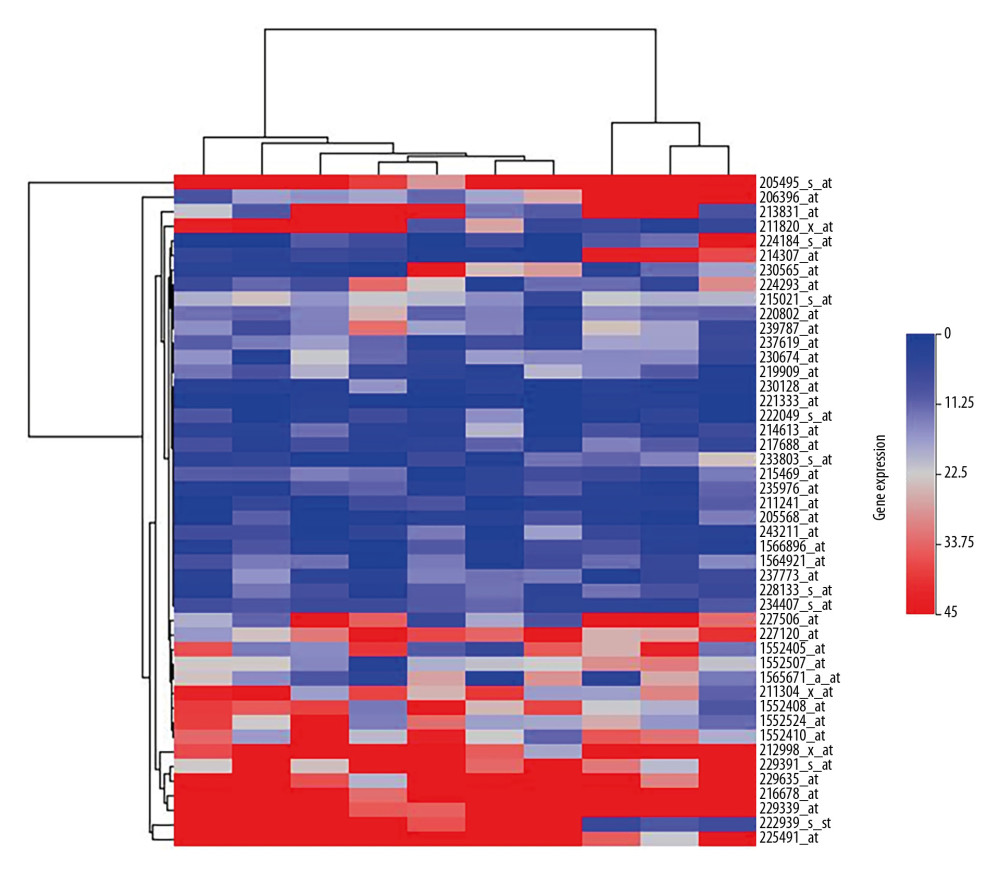 Figure 2. The heatmap of the differentially expressed genes between the Case group and Control group. The expression level was demonstrated by the color range from 0 to 45. The clustered gene signature is shown at the right side of the figure.
Figure 2. The heatmap of the differentially expressed genes between the Case group and Control group. The expression level was demonstrated by the color range from 0 to 45. The clustered gene signature is shown at the right side of the figure. 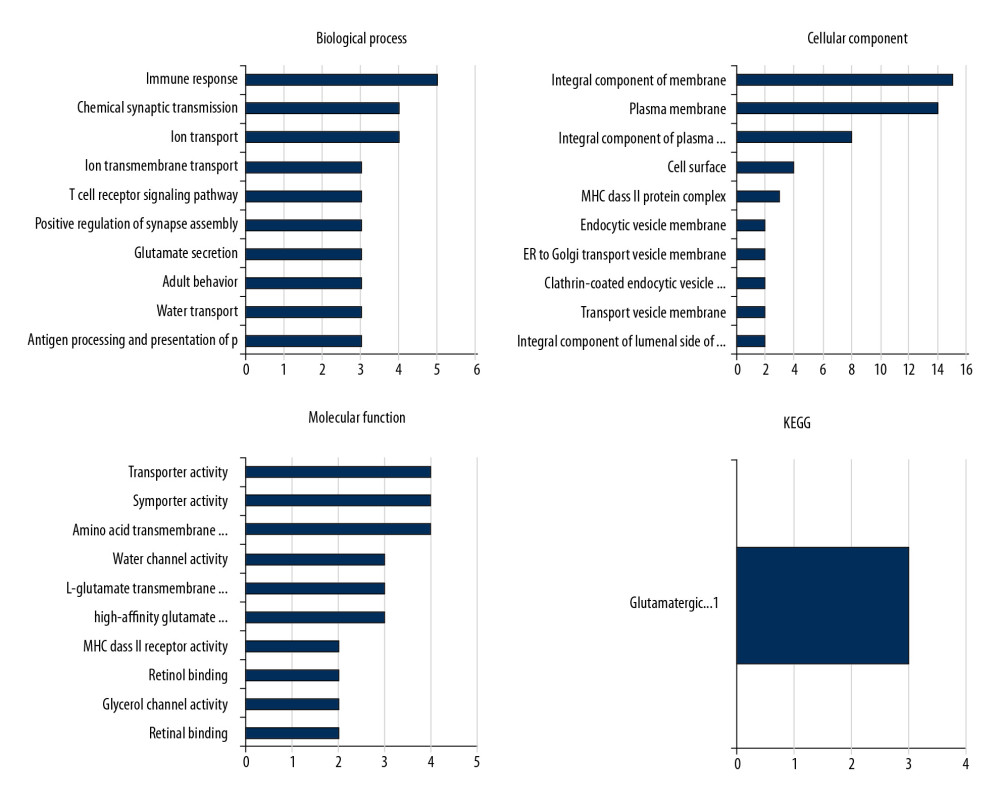 Figure 3. GO and KEGG analysis of the differentially expressed genes. The 46 DEGs were enriched in glutamate secretion, positive regulation of synapse assembly, and T cell receptor signaling pathway. For the molecular function, the genes were included in the aspects of MHC class II receptor activity, high-affinity glutamate transmembrane transporter activity, and L-glutamate trans membrane transporter activity. KEGG pathway analysis indicated the dysregulated genes were only enriched in the glutamatergic synapse pathway.
Figure 3. GO and KEGG analysis of the differentially expressed genes. The 46 DEGs were enriched in glutamate secretion, positive regulation of synapse assembly, and T cell receptor signaling pathway. For the molecular function, the genes were included in the aspects of MHC class II receptor activity, high-affinity glutamate transmembrane transporter activity, and L-glutamate trans membrane transporter activity. KEGG pathway analysis indicated the dysregulated genes were only enriched in the glutamatergic synapse pathway. 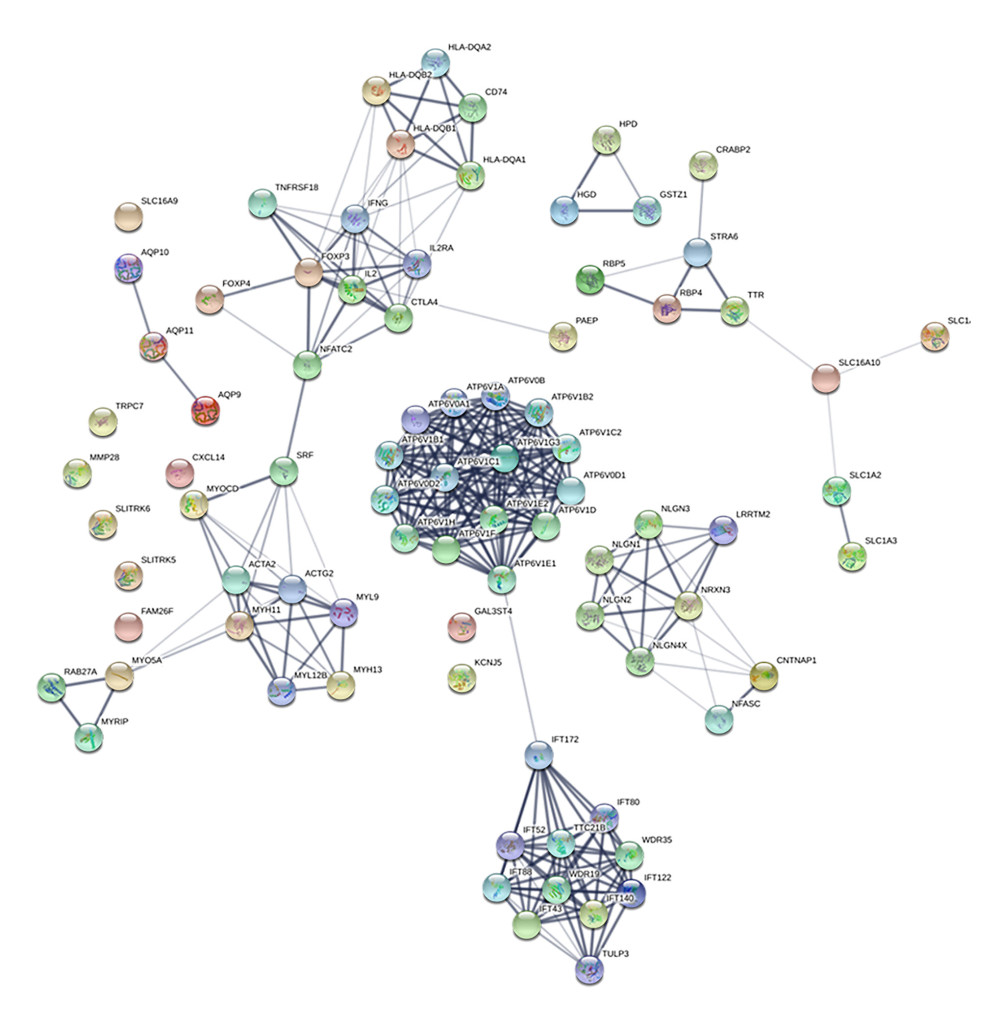 Figure 4. Protein–protein interaction (PPI) network of the dysregulated genes. In the network, 83 nodes and 273 edges with the average node degreed of 6.58 were established.
Figure 4. Protein–protein interaction (PPI) network of the dysregulated genes. In the network, 83 nodes and 273 edges with the average node degreed of 6.58 were established. 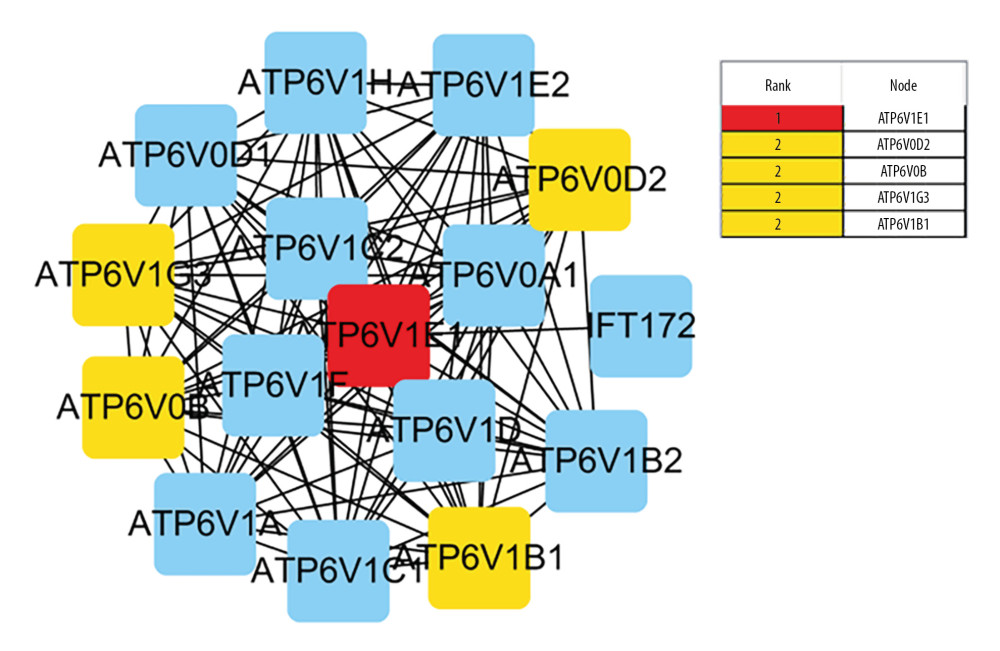 Figure 5. Identified the hub gene by Cytohubba. In the network, ATPTV1E1, ATPTV1E10V1, ATP6V0B, ATP6V1G3, and ATP6V1B1 were calculated as the top 5 hub genes.
Figure 5. Identified the hub gene by Cytohubba. In the network, ATPTV1E1, ATPTV1E10V1, ATP6V0B, ATP6V1G3, and ATP6V1B1 were calculated as the top 5 hub genes. 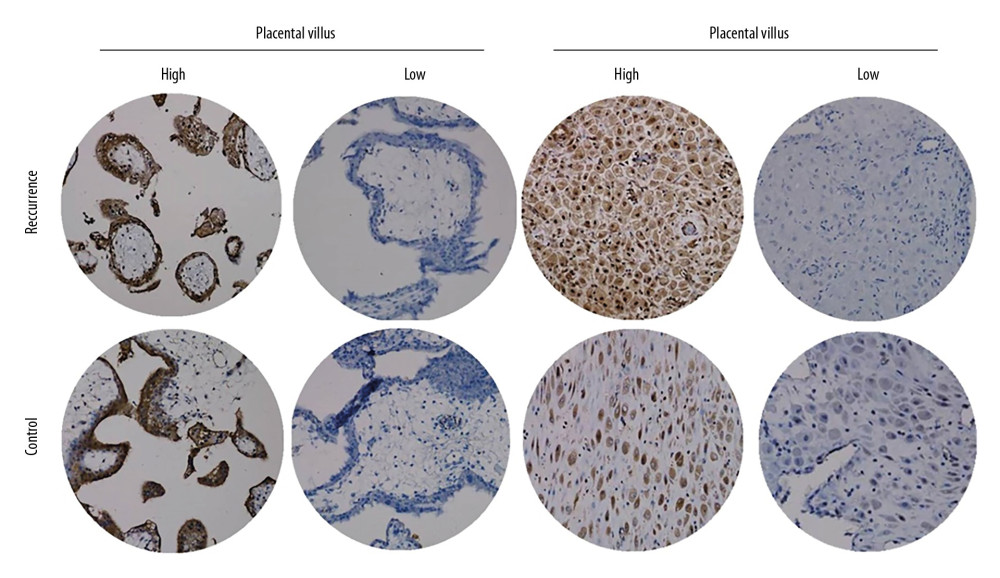 Figure 6. ATP6V1G3 protein expression in placental villus and decidual tissue of placenta of the Case and Control groups detected by immunohistochemistry assay. ATP6V1G3 protein was mainly expressed in the cytoplasm with brownish stain (200×).
Figure 6. ATP6V1G3 protein expression in placental villus and decidual tissue of placenta of the Case and Control groups detected by immunohistochemistry assay. ATP6V1G3 protein was mainly expressed in the cytoplasm with brownish stain (200×). References
1. Naessens A, Foulon W, Cammu H, Epidemiology and pathogenesis of ureaplasma urealyticum in spontaneous abortion and early preterm labor: Acta Obstet Gynecol Scand, 1987; 66; 513-16
2. Osborn J, Spinelli A, Cattaruzza MSTrends of spontaneous abortion in Italy and risk factors: Epidemiol Prev, 2000; 24; 152-53 [in Italian]
3. Schupf N, Ottman R, Reproduction among individuals with idiopathic/cryptogenic epilepsy: Risk factors for spontaneous abortion: Epilepsia, 1997; 38; 824-29
4. Tan NH, Yahya A, Adeeb N, Sociobiological risk factors for spontaneous abortion in Malaysia: J Obstet Gynaecol (Tokyo 1995), 1995; 21; 313-18
5. Domínguez-Rojas V, de Juanes-Pardo JR, Astasio-Arbiza P, Spontaneous abortion in a hospital population: Are tobacco and coffee intake risk factors: Eur J Epidemiol, 1994; 10; 665-68
6. Arjmand F, Ghasemi N, Mirghanizadeh SA, Samadi M, The balance of the immune system between HLA-G and NK cells in unexplained recurrent spontaneous abortion and polymorphisms analysis: Immunol Res, 2016; 64; 785-90
7. Fan X, Wang W, Liu REpidemiological features and risk factors of spontaneous abortion among rural Tibetan women at childbearing age: Zhonghua Liu Xing Bing Xue Za Zhi, 2014; 35; 401-5 [in Chinese]
8. Vaiman D, Genetic regulation of recurrent spontaneous abortion in humans: Biomed J, 2015; 38; 11-24
9. Pereza N, Volk M, Zrakić N, Genetic variation in tissue inhibitors of metalloproteinases as a risk factor for idiopathic recurrent spontaneous abortion: Fertil Steril, 2013; 99; 1923-29
10. Gloria-Bottini F, Nicotra M, Amante A, Adenylate kinase genetic polymorphism and spontaneous abortion: Am J Hum Biol, 2012; 24; 186-88
11. Griffin DK, Larkin DM, O’Connor RE, Time lapse: A glimpse into prehistoric genomics: Eur J Med Genet, 2020; 63(2); 103640
12. Wang K, Wang W, Li M, A brief procedure for big data analysis of gene expression: Animal Model Exp Med, 2018; 1; 189-93
13. Rull K, Tomberg K, Kõks S, Increased placental expression and maternal serum levels of apoptosis-inducing TRAIL in recurrent miscarriage: Placenta, 2013; 34; 141-48
14. Lédée N, Munaut C, Aubert J, Specific and extensive endometrial deregulation is present before conception in IVF/ICSI repeated implantation failures (IF) or recurrent miscarriages: J Pathol, 2011; 225; 554-64
15. Zhou LY, Zhang HX, Lan YL, Epidemiological investigation of risk factors of the pregnant women with early spontaneous abortion in Beijing: Chin J Integr Med, 2017; 23; 345-49
16. Coulam CB, Epidemiology of recurrent spontaneous abortion: Am J Reprod Immunol, 1991; 26; 23-27
17. Bocciolone L, Parazzini F, Fedele LEpidemiology of spontaneous abortion: A review of the literature: Ann Ostet Ginecol Med Perinat, 1989; 110; 323-34 [in Italian]
18. Xu X, Du C, Li H, Association of VEGF genetic polymorphisms with recurrent spontaneous abortion risk: A systematic review and meta-analysis: PLoS One, 2015; 10; e0123696
19. Konovalov OESocial-hygienic and medico-biological risk factors in spontaneous abortion and infertility: Probl Sotsialnoi Gig Istor Med, 1998(1); 19-22
Figures
 Figure 1. Identification of differentially expressed genes. We identified 46 overlapping dysregulated genes through GSE22490 and GSE2678 (A: Volcano plot of GSE22490; B: Volcano plot of GSE26787; C: Venn plot of the overlapping genes).
Figure 1. Identification of differentially expressed genes. We identified 46 overlapping dysregulated genes through GSE22490 and GSE2678 (A: Volcano plot of GSE22490; B: Volcano plot of GSE26787; C: Venn plot of the overlapping genes). Figure 2. The heatmap of the differentially expressed genes between the Case group and Control group. The expression level was demonstrated by the color range from 0 to 45. The clustered gene signature is shown at the right side of the figure.
Figure 2. The heatmap of the differentially expressed genes between the Case group and Control group. The expression level was demonstrated by the color range from 0 to 45. The clustered gene signature is shown at the right side of the figure. Figure 3. GO and KEGG analysis of the differentially expressed genes. The 46 DEGs were enriched in glutamate secretion, positive regulation of synapse assembly, and T cell receptor signaling pathway. For the molecular function, the genes were included in the aspects of MHC class II receptor activity, high-affinity glutamate transmembrane transporter activity, and L-glutamate trans membrane transporter activity. KEGG pathway analysis indicated the dysregulated genes were only enriched in the glutamatergic synapse pathway.
Figure 3. GO and KEGG analysis of the differentially expressed genes. The 46 DEGs were enriched in glutamate secretion, positive regulation of synapse assembly, and T cell receptor signaling pathway. For the molecular function, the genes were included in the aspects of MHC class II receptor activity, high-affinity glutamate transmembrane transporter activity, and L-glutamate trans membrane transporter activity. KEGG pathway analysis indicated the dysregulated genes were only enriched in the glutamatergic synapse pathway. Figure 4. Protein–protein interaction (PPI) network of the dysregulated genes. In the network, 83 nodes and 273 edges with the average node degreed of 6.58 were established.
Figure 4. Protein–protein interaction (PPI) network of the dysregulated genes. In the network, 83 nodes and 273 edges with the average node degreed of 6.58 were established. Figure 5. Identified the hub gene by Cytohubba. In the network, ATPTV1E1, ATPTV1E10V1, ATP6V0B, ATP6V1G3, and ATP6V1B1 were calculated as the top 5 hub genes.
Figure 5. Identified the hub gene by Cytohubba. In the network, ATPTV1E1, ATPTV1E10V1, ATP6V0B, ATP6V1G3, and ATP6V1B1 were calculated as the top 5 hub genes. Figure 6. ATP6V1G3 protein expression in placental villus and decidual tissue of placenta of the Case and Control groups detected by immunohistochemistry assay. ATP6V1G3 protein was mainly expressed in the cytoplasm with brownish stain (200×).
Figure 6. ATP6V1G3 protein expression in placental villus and decidual tissue of placenta of the Case and Control groups detected by immunohistochemistry assay. ATP6V1G3 protein was mainly expressed in the cytoplasm with brownish stain (200×). In Press
05 Mar 2024 : Clinical Research
Role of Critical Shoulder Angle in Degenerative Type Rotator Cuff Tears: A Turkish Cohort StudyMed Sci Monit In Press; DOI: 10.12659/MSM.943703
06 Mar 2024 : Clinical Research
Comparison of Outcomes between Single-Level and Double-Level Corpectomy in Thoracolumbar Reconstruction: A ...Med Sci Monit In Press; DOI: 10.12659/MSM.943797
21 Mar 2024 : Meta-Analysis
Economic Evaluation of COVID-19 Screening Tests and Surveillance Strategies in Low-Income, Middle-Income, a...Med Sci Monit In Press; DOI: 10.12659/MSM.943863
10 Apr 2024 : Clinical Research
Predicting Acute Cardiovascular Complications in COVID-19: Insights from a Specialized Cardiac Referral Dep...Med Sci Monit In Press; DOI: 10.12659/MSM.942612
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952