12 May 2021: Clinical Research
Performance of Diagnostic Model for Differentiating Between COVID-19 and Influenza: A 2-Center Retrospective Study
Jingwen Li1E, Simin Li2B, Xiaoming Qiu3B, Wenyan Zhu2F, Linfeng Li2F, Bo Qin4A*DOI: 10.12659/MSM.932361
Med Sci Monit 2021; 27:e932361
Abstract
BACKGROUND: COVID-19 and influenza share many similarities, such as mode of transmission and clinical symptoms. Failure to distinguish the 2 diseases may increase the risk of transmission. A fast and convenient differential diagnosis between COVID-19 and influenza has significant clinical value, especially for low- and middle-income countries with a shortage of nucleic acid detection kits. We aimed to establish a diagnostic model to differentiate COVID-19 and influenza based on clinical data.
MATERIAL AND METHODS: A total of 493 patients were enrolled in the study, including 282 with COVID-19 and 211 with influenza. All data were collected and reviewed retrospectively. The clinical and laboratory characteristics of all patients were analyzed and compared. We then randomly divided all patients into development sets and validation sets to establish a diagnostic model using multivariate logistic regression analysis. Finally, we validated the diagnostic model using the validation set.
RESULTS: We preliminarily established a diagnostic model for differentiating COVID-19 from influenza that consisted of 5 variables: age, dry cough, fever, white cell count, and D-dimer. The model showed good performance for differential diagnosis.
CONCLUSIONS: This initial model including clinical features and laboratory indices effectively differentiated COVID-19 from influenza. Patients with a high score were at a high risk of having COVID-19, while patients with a low score were at a high risk of having influenza. This model could help clinicians quickly identify and isolate cases in the absence of nucleic acid tests, especially during the cocirculation of COVID-19 and influenza. Owing to the study’s retrospective nature, further prospective study is needed to validate the accuracy of the model.
Keywords: COVID-19, Influenza, Human, Models, Statistical, COVID-19, Cough, Diagnosis, Differential, Fever, SARS-CoV-2
Background
The COVID-19 pandemic is on-going [1]. The strong infectivity and the unknown characteristics of this disease impose a tremendous burden on health systems worldwide [2]. The availability of SARS-CoV-2 vaccine in several countries brings hope for ending the pandemic [3]. The key for the control and prevention of COVID-19 will be transferred from high-income countries to low- and middle-income countries (LMICs) due to global inequity in COVID-19 vaccination. For LMICs, reducing the number of new cases of COVID-19 is crucial for the prevention and control of the pandemic. Affordable, effective, and fast diagnosis is still lacking [4].
When COVID-19 emerged, influenza was still widespread [5]. Influenza is the most common respiratory tract disease in the autumn and winter [6], and it can be transmitted via the respiratory route and through direct contact [7]. The majority of patients present with mild-to-moderate respiratory symptoms, but some may progress to severe complications [8]. COVID-19 and influenza share many similarities including transmission and clinical manifestations such as fever, dry cough, sore throat, headache, myalgia, and weakness [9,10], and these similarities may easily lead to misdiagnosis of the 2 diseases. For example, some patients who had influenza-like symptoms had positive test results for SARS-CoV-2 in a study at the Los Angeles Medical Center [11]. Failure to distinguish patients with COVID-19 from those with influenza may significantly increase the risk of cross-infection and promote COVID-19 transmission. In addition, treatments for COVID-19 and influenza are very different, and misdiagnosis could result in treatment delay and unnecessary wastage of medical resources. Rapid differential diagnosis between COVID-19 and influenza remains a great challenge and is important for the next phase of outbreak control.
Nucleic acid detection is the diagnostic criterion standard for COVID-19 and influenza infection [12,13]. However, it requires expensive reagents, such as RNA extraction kits, and quantitative polymerase chain reaction (PCR) machines [13]. The costs of nucleic acid detection and rapid molecular assays are unaffordable for LMICs. Rapid molecular assays (nucleic acid amplification tests) and rapid influenza diagnostic tests are priorities for influenza diagnosis, which may be insufficient for those countries and areas [14,15]. Meanwhile, the COVID-19 pandemic causes economic devastation and worsens poverty [16]. Thus, the pandemic trend in poorer countries is unclear and unpredictable due to the shortage of nucleic acid detection kits and critical medical equipment [17–19]. A complementary method for these preferred diagnostic approaches is needed, and our diagnostic model based on clinical data can be used as a rapid and inexpensive method.
Differential diagnostic models based on clinical features and routine laboratory testing have important clinical significance for pneumonia classification. Sampedro et al [20] created a model based on electronic health records that could predict COVID-19 severity and demonstrated a fair to good fit. Formica et al [21] used 8 complete blood count predictors and established a scoring system for COVID-19 with a discriminatory power of 88% based on the area under the receiver-operating characteristic (ROC) curve (AUC). This system waives the need for swab tests for asymptomatic patients with a low score. A model combining clinical features and inflammatory markers has proven to be useful for predicting the bacterial etiology of community-acquired pneumonia, with a specificity of 96.6% [22,23]. Several biomarkers such as monocyte count, percentage of basophils, and time from symptom onset to hospital admission were significantly different between COVID-19 and influenza, which could help the differential diagnosis [24–29]. These investigations were helpful in the differential diagnosis and elucidated the differing pathogenesis of the 2 diseases. However, some studies were single-center studies with relatively small sample sizes or were based on single-dimension data such as biochemical indicators or clinical features [26,27]. Also, patients involved in another 2 studies were recruited from the Emergency Department, leading to selection bias [24,25]. Larger sample sizes, multidimensional data, and more representative patients are needed for a definitive conclusion.
The purpose of the current study was to build a clinical diagnostic model that can differentiate COVID-19 and influenza. This model was based on multidimensional clinical data from 2 hospitals, and it was intended to serve as a fast-screening method to ease the diagnostic difficulties due to insufficient medical supplies and help physicians quickly determine different isolation measures.
Material and Methods
ETHICS:
The study was performed according to the principles of the Declaration of Helsinki and was approved by the First Affiliated Hospital to Army Medical University Ethics Committee (Approval No. KY2020036).
STUDY DESIGN AND PATIENTS:
This study was designed to build a diagnostic model for differentiating between COVID-19 and influenza. The model was based on epidemiology and the clinical features of hospitalized patients with COVID-19 and influenza. All patients came from 2 hospitals under supervision of China National Health Commission (NHC), including Huangshi Central Hospital, Affiliated Hospital of Hubei Polytechnic University (from January 2019 to March 2020), and the First Affiliated Hospital of Army Medical University (from January 2019 to March 2020). These hospitals are large university teaching hospitals in the southwest region of China for the population of that area. A total of 493 eligible patients were enrolled in the study, including 282 COVID-19 patients and 211 influenza patients.
SELECTION CRITERIA:
Inclusion criteria for the study were adult patients diagnosed with COVID-19 or influenza. COVID-19 was diagnosed according to the Diagnosis and Treatment Protocol for COVID-19 by the NHC of the People’s Republic of China (trial seventh edition) [30], and the diagnosis of influenza was based on Diagnosis and Treatment Protocol for Influenza from the NHC of China [31]. All the enrolled patients with COVID-19 had positive SARS-CoV-2 PCR results, negative PCR results for influenza, and negative test results for other respiratory viral infections. Influenza patients had positive PCR results for the influenza virus or positive influenza antigen detection, as well as negative results for other respiratory viral infections. Exclusion criteria included patients younger than 18 years of age, pregnant or lactating women, patients with other respiratory infections such as parainfluenza and respiratory syncytial virus, and patients with incomplete clinical data.
DATA COLLECTION:
Medical information including demographic data, clinical features, and laboratory test results were collected from medical records and reviewed respectively. Respiratory specimens from patients were collected to identify the SARS-CoV-2 infection [32]. Respiratory specimens or blood samples were collected to identify influenza infection using PCR tests or influenza antigen detection [33]. All symptoms and laboratory data were collected simultaneously with pathogen or antibody tests on the first day of hospital admission unless indicated otherwise.
STATISTICAL ANALYSIS:
The COVID-19 and influenza groups were each randomized into development and validation sets respectively. The development sets were used to establish a diagnostic model, and the validation sets were used to evaluate it. Based on the development set data, all the statistically significant variables (
Results
DEMOGRAPHIC AND CLINICAL FEATURES:
As shown in Table 1, a total of 493 eligible patients were enrolled, including 282 (57%) COVID-19 patients and 211 (43%) influenza patients. Numerous differences were identified between the COVID-19 group and the influenza group. Patients with COVID-19 were significantly older (mean age, 49.6±14.4 years vs 42.7±16.4 years; P<0.001). A history of hypertension (COVID-19: 9.9% vs influenza: 22.0%, P<0.001), diabetes mellitus (COVID-19: 7.1% vs influenza: 17.5%, P<0.001), and cardiovascular disease (COVID-19: 8.2% vs influenza: 13.7%, P=0.045) were significantly different between the 2 diseases. In addition, the COVID-19 patients were more prone than the influenza patients to present fever (209 [74.1%] vs 101 [47.9%]; P<0.001), dry cough (193 [68.4%] vs 64 [30.3%]; P<0.001), fatigue (86 [30.5%] vs 10 [4.7%]; P<0.001), myalgia (24 [8.5%] vs 2 [0.9%]; P<0.001), and dyspnea (19 [6.7%] vs 5 [2.4%]; P=0.033). However, the patients with influenza more often experienced faster pulses (87±12 beats/min vs 96±17 beats/min; P<0.001). The mean time from illness onset to hospital admission in COVID-19 patients was shorter than the median time in influenza patients (11.9±5.3 days vs 15.2±4.4 days; P=0.024)
LABORATORY FEATURES:
As with the epidemiological characteristics, many differences in laboratory tests were also found between patients with COVID-19 and those with influenza (Table 2). In terms of routine blood test results, decreased levels of white blood cells (WBCs) ([5.2±2.2]×109/L vs [7.6±3.9]×109/L; P<0.001) and neutrophils ([3.5±2.1]×109/L vs [5.6±3.7]×109/L; P<0.001) were identified in the COVID-19 patients. In addition, lower levels of prothrombin time (11.8±2.6 s vs 12.5±2.4 s; P=0.020) and D-dimer (0.4±0.9 mg/L vs 1.8±1.2 mg/L; P<0.001) were found in the COVID-19 group compared with the influenza group. Moreover, decreased concentrations of alanine aminotransferase (AST) (P=0.005), aspartate aminotransferase (ALT) (P=0.001), bilirubin (P<0.001), C-reactive protein (P<0.001), and creatinine (P=0.005) were confirmed in COVID-19 patients; the influenza patients showed opposite trends. However, the influenza group presented lower concentrations of potassium (P<0.001) and sodium (P=0.003), and the activated partial thromboplastin time (aPTT) was also reduced (P<0.001). No obvious differences were found regarding other indicators.
DIAGNOSTIC MODEL FOR DISTINGUISHING BETWEEN COVID-19 AND INFLUENZA:
To establish a diagnostic model, we randomly assigned COVID-19 patients (n=282) to a development set (n=226) and a validation set (n=56). Influenza patients (n=211) were also split into a development set (n=168) and a validation set (n=43). The mean age (46.6±15.6 years vs 46.8±15.8 years,
All the statistically significant variables (
Codes used for the equations:
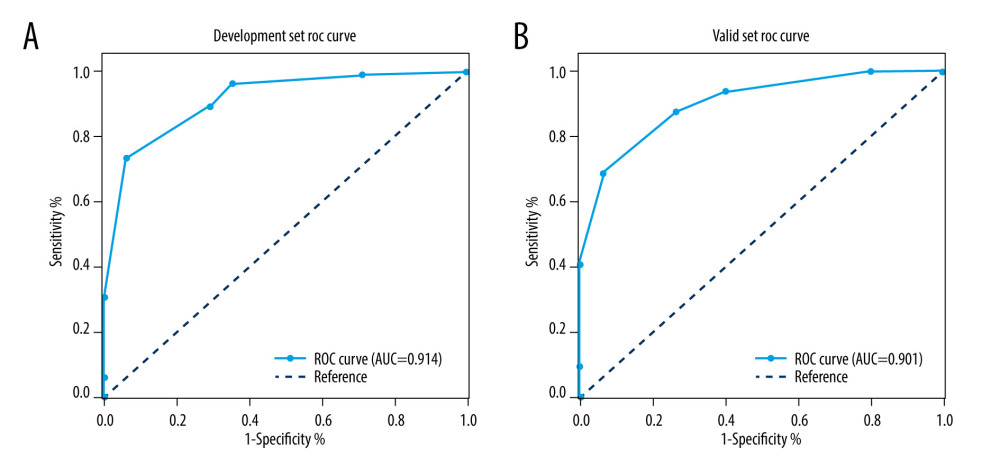
Dry cough, age, and fever were distinguishing factors of COVID-19, whereas D-dimer and WBC count were distinguishing factors of influenza. ROC analysis indicated the AUC of the model was 0.914 (95% confidence interval [CI], 0.882–0.948) (Figure 1A). When the cutoff value was 0.36, the sensitivity, specificity, and accuracy were 70.8%, 89.4%, and 83.9%, respectively (Table 3).
VALIDATION OF THE DIAGNOSTIC MODEL:
We further validated the diagnostic model based on the validation data set. As shown in Figure 1, ROC analysis showed the AUC of the model for the validation set was 0.901 (95% CI=0.819–0.971), and there were no striking differences in AUC between the development set (0.914, 95% CI=0.882–0.948) and validation set (0.901, 95% CI=0.819–0.971; P=0.112). Based on the ROC analysis, the diagnostic model had good distinction power in the 2 data sets (Figure 1). When the cutoff value was 0.36, the sensitivity, specificity, positive predictive value, negative predictive value, and accuracy of the validation set were 0.733, 0.875, 0.875, 0.733, and 0.830, respectively (Table 3).
Discussion
In this study, we developed a COVID-19 predictive model based on multidimensional medical data, including epidemiological history, clinical symptoms, and laboratory indices from 2 hospitals. We then verified the accuracy of the model using a validation set. Five markers that were critical for differentiating the 2 diseases were identified, including age, dry cough, fever, WBC count, and D-dimer. We also found other indicators that may have a certain significance for differential diagnosis such as AST, aPTT, fatigue, headache, creatinine, and dyspnea. These abnormalities revealed that the damage of SARS-CoV-2 infection may involve cellular immune response, coagulation disorder, and organ injury. It is also believed that a cytokine storm may play an important role [34].
Numerous methods have been developed for COVID-19 diagnosis. Among these, whole-genome sequencing was reported to have high specificity and sensitivity, but the high cost and time-consuming nature limited clinical application [12]. Single-cell sequencing was also used to distinguish between the 2 diseases. Liu et al [35] reported the single-cell transcriptional landscape of peripheral blood mononuclear cells in COVID-19 patients and influenza A virus patients; it showed distinct immune response pathways in the 2 diseases, but this method currently is limited to scientific research. The utility of clinical signs, symptoms, and serum biomarkers could discriminate bacterial from viral pneumonia with a sensitivity of 75% and a specificity of 84% [36]. Although this approach was accurate, it was not applied in community hospitals owing to the need for large computational resources. By being fast and simple, our model is suitable for community hospitals and LMICs with shortages of pathogenic detection kits and critical medical equipment, especially during the pandemic.
Our study built this predictive model for differentiating COIVID-19 and influenza. In a previous study, Zhang et al [27] compared peripheral blood parameters of COVID-19 patients and influenza patients and found some differences, such as older age and reduced WBC counts for COVID-19 patients, which were consistent with our study. One study by Sieber et al [24] also contrasted the 2 categories of patients from the Emergency Department and showed that the most important difference was a longer latency from symptom onset to hospital admission for COVID-19 patients. This was different from our study, and this difference could be attributable to the studies having different study populations. COVID-19 prevention and control strategies varied between countries. The strict quarantine measures and screening policies in China ensured that COVID-19 patients could get the earliest identification and medical treatments during the pandemic, which made the time from symptom onset to hospital admission shorter. Alternatively, some differences that exist among the current studies may be due to the use of different sample sizes [25,26]. Future studies with larger sample sizes could be done to validate the results of this study. Although the previous studies presented the epidemiological characteristics, clinical features, and laboratory indicators for COVID-19 and influenza, they did not build a comprehensive predictive model from those indices. The noncomprehensiveness of these indicators resulted in low diagnostic performance, and results could not be applied in clinical practice. Jung et al [37] utilized a similar computational approach to develop a diagnostic model to effectively differentiate Crohn disease from intestinal tuberculosis [37]. We hope our model established by the use of this method offers a reliable and practical approach to distinguish COVID-19 and influenza.
We found that age was an important factor in distinguishing patients with COVID-19 from those with influenza. COVID-19 patients were older than influenza patients. The influenza A virus predominantly affects children and young adults more than older adults, and although the death rates are highest in the >65 years group, overall, more deaths are reported in younger people [38–40]. Influenza B viruses also tend to preferentially affect children and young adults, and significant mortality has been confirmed, particularly in the pediatric population [41,42]. The possible reasons for this pattern are that older patients may have a cross-reactive antibody that confers protection to the current influenza strain [38]. According to the recent largest sample study of COVID-19, most patients were 30 to 79 years old (87%) and 3% were 80 or older [43]. A joint investigation report on COVID-19 released by the World Health Organization on February 29, 2020, revealed that the median age of infected patients was 51 years, and 78% of patients were between 30 and 69 years [44]. The data demonstrated that chronic disease, a weak immune system, and poor overall health may increase the susceptibility to SARS-CoV-2 infection.
Fever was one of the markers in the predictive model. Patients with COVID-19 were more prone to have fever compared with those with influenza, which was consistent with a previous study [25]. According to Zhong et al [45], about half of the patients had fever on initial presentation and fever developed in 87.9% of patients following hospitalization. We speculated that the majority of COVID-19 patients were older and had chronic comorbidities and weakened immunity, the virus replicated rapidly in vivo and activated the immune system, and then fever occurred. Previous findings support our hypothesis [46,47]. In the early stage of COVID-19, a normal or decreased total WBC count was a characteristic of viral infection [48]. Compared with influenza, the WBC count being lower with COVID-19 may be due to immune system damage and cytokine storm.
Our study has some limitations. First, studies with expanded samples and prospective clinical trials are required for further validation and higher accuracy. Second, the patients in this study came from the southwest region of China, and the results need to be interpreted with caution. Third, all patients enrolled in this study were inpatients rather than outpatients, which may lead to selection bias. Our model would be more applicable to inpatients.
Conclusions
We built a diagnostic model for distinguishing COVID-19 from influenza. The markers for distinguishing COVID-19 from influenza were age, fever, dry cough, WBC count, and D-dimer. This model will assist clinicians in distinguishing the 2 diseases and accelerate diagnosis and quarantine, and it can even serve as a fast-screening method in community hospitals and LMICs with insufficient testing supplies. All findings should be interpreted with caution, and this model should be validated in larger prospective cohort studies.
References
1. The Lancet, Science during COVID-19: where do we go from here?: Lancet, 2021; 396(10267); 1941
2. Kamel Boulos MN, Geraghty EM: Int J Health Geogr, 2020; 19(1); 8
3. Mishra SK, Tripathi T, One year update on the COVID-19 pandemic: Where are we now?: Acta Trop, 2021; 214; 105778
4. Castillo JC, Ahuja A, Athey S, Market design to accelerate COVID-19 vaccine supply: Science, 2021; 371(6534); 1107-9
5. Rubin R, What happens when COVID-19 collides with flu season?: JAMA, 2020; 324(10); 923-25
6. Labella AM, Merel SE, Influenza: Med Clin North Am, 2013; 97(4); 621-45
7. Chow EJ, Doyle JD, Uyeki TM, Influenza virus-related critical illness: prevention, diagnosis, treatment: Crit Care, 2019; 23(1); 214
8. Iuliano AD, Roguski KM, Chang HH, Estimates of global seasonal influenza-associated respiratory mortality: A modelling study: Lancet, 2018; 391(10127); 1285-300
9. Belongia EA, Osterholm MT, COVID-19 and flu, a perfect storm: Science, 2020; 368(6496); 1163
10. Singer BD, COVID-19 and the next influenza season: Sci Adv, 2020; 6(31); eabd0086
11. Spellberg B, Haddix M, Lee R, Community prevalence of SARS-CoV-2 among patients with influenzalike illnesses presenting to a Los Angeles medical center in March 2020: JAMA, 2020; 323(19); 1966-67
12. Udugama B, Kadhiresan P, Kozlowski HN, Diagnosing COVID-19: the disease and tools for detection: ACS Nano, 2020; 14(4); 3822-35
13. Esbin MN, Whitney ON, Chong S, Overcoming the bottleneck to widespread testing: A rapid review of nucleic acid testing approaches for COVID-19 detection: RNA (New York, NY), 2020; 26(7); 771-83
14. Uyeki TM, Bernstein HH, Bradley JS, Clinical practice guidelines by the Infectious Diseases Society of America: 2018 update on diagnosis, treatment, chemoprophylaxis, and institutional outbreak management of seasonal influenzaa: Clin Infect Dis, 2019; 68(6); 895-902
15. Idubor OI, Kobayashi M, Ndegwa L, Improving detection and response to respiratory events – Kenya, April 2016–April 2020: MMWR Morb Mortal Wkly Rep, 2020; 69(18); 540-44
16. Gupta M, Abdelmaksoud A, Jafferany M, COVID-19 and economy: Dermatol Ther, 2020; 33(4); e13329
17. Moreno-Contreras J, Espinoza MA, Sandoval-Jaime C, Saliva sampling and its direct lysis, an excellent option to increase the number of SARS-CoV-2 diagnostic tests in settings with supply shortages: J Clin Microbiol, 2020; 58(10); e01659-20
18. Hopman J, Allegranzi B, Mehtar S, Managing COVID-19 in low- and middle-income countries: JAMA, 2020; 323(16); 1549-50
19. Lloyd-Sherlock P, Ebrahim S, Geffen L, McKee M, Bearing the brunt of covid-19: Older people in low and middle income countries: BMJ, 2020; 368; m1052
20. Gude-Sampedro F, Fernández-Merino C, Ferreiro L, Development and validation of a prognostic model based on comorbidities to predict COVID-19 severity: A population-based study: Int J Epidemiol, 2020; 50(1); 64-74
21. Formica V, Minieri M, Bernardini S, Complete blood count might help to identify subjects with high probability of testing positive to SARS-CoV-2: Clin Med (Lond), 2020; 20(4); e114-19
22. Alcoba G, Keitel K, Maspoli V, A three-step diagnosis of pediatric pneumonia at the emergency department using clinical predictors, C-reactive protein, and pneumococcal PCR: Eur J Pediatr, 2017; 176(6); 815-24
23. Berg AS, Inchley CS, Fjaerli HO, Clinical features and inflammatory markers in pediatric pneumonia: A prospective study: Eur J Pediatr, 2017; 176(5); 629-38
24. Sieber P, Flury D, Güsewell S, Characteristics of patients with coronavirus disease 2019 (COVID-19) and seasonal influenza at time of hospital admission: A single center comparative study: BMC Infect Dis, 2021; 21(1); 271
25. Bouzid D, Mullaert J, Le Hingrat Q, Characteristics associated with COVID-19 or other respiratory viruses’ infections at a single-center emergency department: PLoS One, 2020; 15(12); e0243261
26. Zayet S, Kadiane-Oussou NJ, Lepiller Q, Clinical features of COVID-19 and influenza: A comparative study on Nord Franche-Comte cluster: Microbes Infect, 2020; 22(9); 481-88
27. Chen J, Pan Y, Li G, Distinguishing between COVID-19 and influenza during the early stages by measurement of peripheral blood parameters: J Med Virol, 2021; 93(2); 1029-37
28. Auvinen R, Nohynek H, Syrjanen R, Comparison of the clinical characteristics and outcomes of hospitalized adult COVID-19 and influenza patients – a prospective observational study: Infect Dis, 2021; 53(2); 111-21
29. Kazancioglu S, Bastug A, Ozbay BO, The role of haematological parameters in patients with COVID-19 and influenza virus infection: Epidemiol Infect, 2020; 148; e272
30. China National Health Commission. The National Health Commission of China: New coronavirus pneumonia prevention and control program, 2020, General Office of the National Health Commission
31. China National Health Commission. The National Health Commission of China: Diagnosis and treatment of influenza (2020), 2020, General Office of the National Health Commission
32. Liu R, Han H, Liu F, Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020: Clin Chim Acta, 2020; 505; 172-75
33. Cifu A, Levinson W, Influenza: JAMA, 2000; 284(22); 2847-49
34. Channappanavar R, Perlman S, Pathogenic human coronavirus infections: Causes and consequences of cytokine storm and immunopathology: Semin Immunopathol, 2017; 39(5); 529-39
35. Zhu L, Yang P, Zhao Y, Single-cell sequencing of peripheral mononuclear cells reveals distinct immune response landscapes of COVID-19 and influenza patients: Immunity, 2020; 53(3); 685-96.e3
36. Bhuiyan MU, Blyth CC, West R, Combination of clinical symptoms and blood biomarkers can improve discrimination between bacterial or viral community-acquired pneumonia in children: BMC Pulm Med, 2019; 19(1); 71
37. Jung Y, Hwangbo Y, Yoon SM, Predictive factors for differentiating between Crohn’s disease and intestinal tuberculosis in Koreans: Am J Gastroenterol, 2016; 111(8); 1156-64
38. Singanayagam A, Singanayagam A, Wood V, Chalmers JD, Factors associated with severe illness in pandemic 2009 influenza A (H1N1) infection: Implications for triage in primary and secondary care: J Infect, 2011; 63(4); 243-51
39. Maruyama T, Fujisawa T, Suga S, Outcomes and prognostic features of patients with influenza requiring hospitalization and receiving early antiviral therapy: A prospective multicenter cohort study: Chest, 2016; 149(2); 526-34
40. Campbell A, Rodin R, Kropp R, Risk of severe outcomes among patients admitted to hospital with pandemic (H1N1) influenza: CMAJ, 2010; 182(4); 349-55
41. Tran D, Vaudry W, Moore D, Hospitalization for influenza A versus B: Pediatrics, 2016; 138(3); e20154643
42. Gavigan P, McCullers JA, Influenza: Annual seasonal severity: Curr Opin Pediatr, 2019; 31(1); 1121-18
43. Wu Z, McGoogan JM, Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: Summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention: JAMA, 2020; 323(13); 1239-42
44. World Health Organization (WHO): Report of the WHO-China Joint Mission on Coronavirus Disease 2019 (COVID-19) [Internet], 2020, Geneva, WHO https://www.who.int/docs/default-source/coronaviruse/who-china-joint-mission-on-covid-19-final-report.pdf
45. Guan W-j, Ni Z-y, Hu Y, Clinical characteristics of 2019 novel coronavirus infection in China: N Engl J Med, 2020; 382(18); 1708-20
46. Gu J, Gong E, Zhang B, Multiple organ infection and the pathogenesis of SARS: J Exp Med, 2005; 202(3); 415-24
47. Kim KD, Zhao J, Auh S, Adaptive immune cells temper initial innate responses: Nat Med, 2007; 13(10); 1248-52
48. Han R, Huang L, Jiang H, Early clinical and CT manifestations of coronavirus disease 2019 (COVID-19) pneumonia: Am J Roentgenol, 2020; 215(2); 338-43
Tables
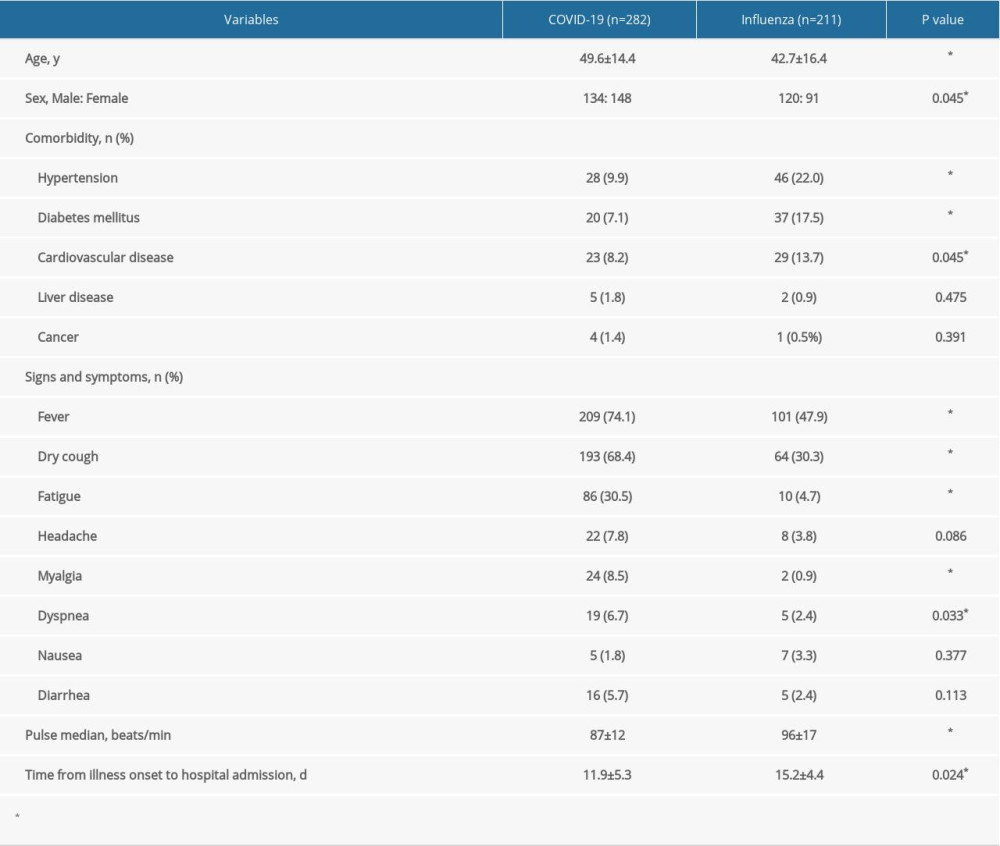 Table 1. Demographic features of patients with COVID-19 and influenza.
Table 1. Demographic features of patients with COVID-19 and influenza.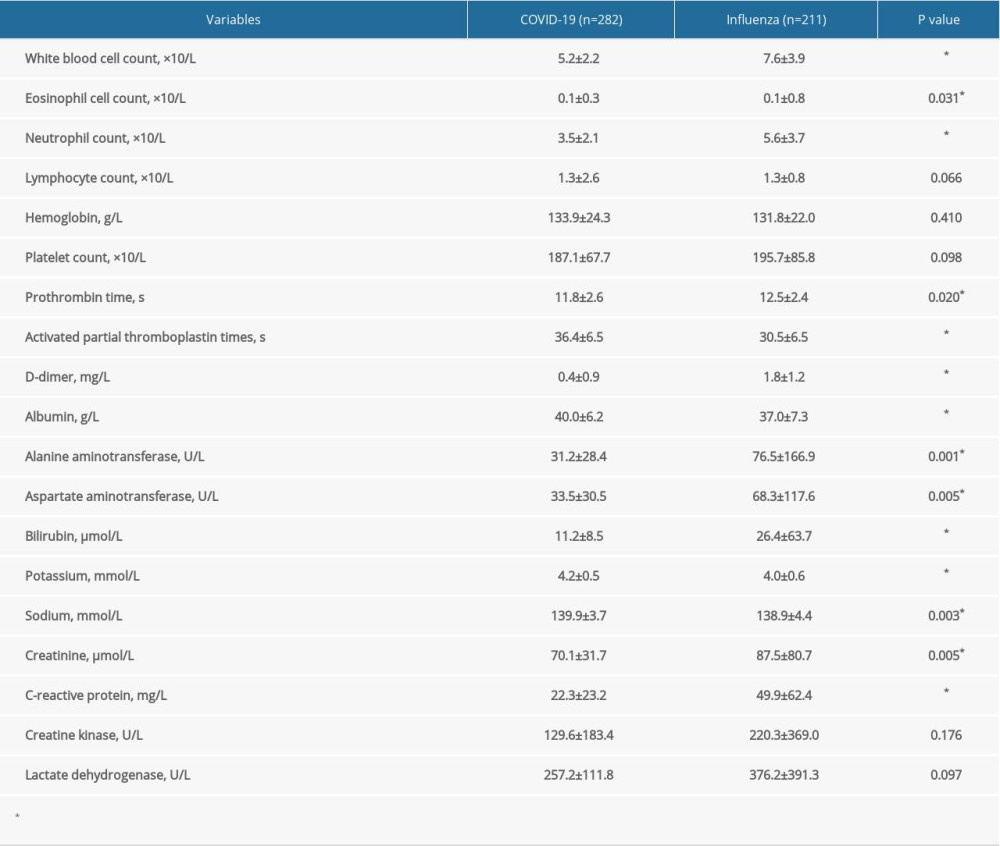 Table 2. Initial laboratory indices of patients with COVID-19 and influenza.
Table 2. Initial laboratory indices of patients with COVID-19 and influenza.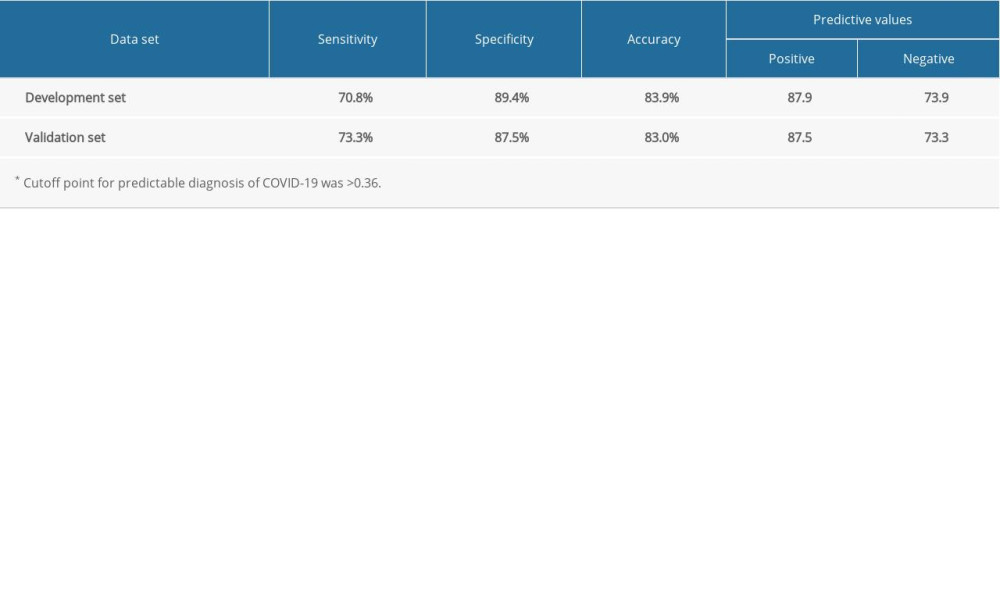 Table 3. Sensitivity, specificity, accuracy, and predictive values of the 5-marker COVID-19 risk score by data set*.
Table 3. Sensitivity, specificity, accuracy, and predictive values of the 5-marker COVID-19 risk score by data set*. Table 1. Demographic features of patients with COVID-19 and influenza.
Table 1. Demographic features of patients with COVID-19 and influenza. Table 2. Initial laboratory indices of patients with COVID-19 and influenza.
Table 2. Initial laboratory indices of patients with COVID-19 and influenza. Table 3. Sensitivity, specificity, accuracy, and predictive values of the 5-marker COVID-19 risk score by data set*.
Table 3. Sensitivity, specificity, accuracy, and predictive values of the 5-marker COVID-19 risk score by data set*. In Press
05 Mar 2024 : Clinical Research
Muscular Function Recovery from General Anesthesia in 132 Patients Undergoing Surgery with Acceleromyograph...Med Sci Monit In Press; DOI: 10.12659/MSM.942780
05 Mar 2024 : Clinical Research
Effects of Thermal Insulation on Recovery and Comfort of Patients Undergoing Holmium Laser LithotripsyMed Sci Monit In Press; DOI: 10.12659/MSM.942836
05 Mar 2024 : Clinical Research
Role of Critical Shoulder Angle in Degenerative Type Rotator Cuff Tears: A Turkish Cohort StudyMed Sci Monit In Press; DOI: 10.12659/MSM.943703
06 Mar 2024 : Clinical Research
Comparison of Outcomes between Single-Level and Double-Level Corpectomy in Thoracolumbar Reconstruction: A ...Med Sci Monit In Press; DOI: 10.12659/MSM.943797
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952