17 February 2023: Clinical Research
EXOC3L1: A Novel Prognostic Biomarker Correlated with Immune Infiltration in Esophageal Squamous Cell Carcinoma
Zhan-Fei Zhang12ABCDEFG*DOI: 10.12659/MSM.938512
Med Sci Monit 2023; 29:e938512
Abstract
BACKGROUND: Exocyst complex component 3-like 1 (EXOC3L1) is ubiquitously present in multiple organs. However, its role in esophageal squamous cell carcinoma (ESCC) remains unknown. The aim of this study was to explore the relationship between EXOC3L1 and ESCC.
MATERIAL AND METHODS: A total of 652 normal samples and 82 ESCC samples obtained from the University of California Santa Cruz (UCSC) Xena were applied to detect the expression difference of EXOC3L1. GSE53625 with 179 paired samples and GSE161533 with 28 paired samples were used for validation. The correlation between clinicopathological features and EXOC3L1 expression was calculated. Kaplan-Meier method was employed to assess the prognostic value of EXOC3L1 in ESCC. Univariate and multivariate Cox regression analyses were carried out to screen the factors contributing to the prognosis of ESCC. In addition, functional enrichment analysis, protein–protein interaction (PPI) network analysis, and immune infiltration analysis were conducted to identify the significantly involved functions of EXOC3L1.
RESULTS: EXOC3L1 was significantly overexpressed in ESCC compared to normal samples. High expression of EXOC3L1 was associated with worse prognosis, and univariate and multivariate Cox regression analysis demonstrated that EXOC3L1 was an independent prognostic predictor of ESCC. Functional enrichment analysis and immune infiltration analysis disclosed that the expression of EXOC3L1 was correlated with the abundance of several types of immune cells.
CONCLUSIONS: EXOC3L1 plays a crucial role in the prognosis of ESCC, and it may serve as a reliable biomarker for predicting the survival and a potential therapeutic target for ESCC.
Keywords: Esophageal Neoplasms, EXOC3 protein, human, Prognosis, tumor microenvironment, Humans, esophageal squamous cell carcinoma, Multivariate Analysis, Protein Interaction Maps
Background
Esophageal carcinoma (EC) is the eighth most common cancer and the sixth most common cause of cancer-related mortality in the world [1], including esophageal adenocarcinoma (EAC) and esophageal squamous cell carcinoma (ESCC) [2,3]. China accounts for more than half of the world’s EC patients and more than 90% of those are ESCC [4,5]. Despite the developments in multi-disciplinary team therapy strategies, such as surgery, chemotherapy, radiotherapy, and immunotherapy, the 5-year overall survival (OS) rate remains unsatisfactory, being less than 30% [6–8], even in developed countries [9]. The low survival rate of ESCC is partly due to the lack of effective therapeutic targets [10]. Although many biomarkers have been explored in ESCC, such as epidermal growth factor receptor (EGFR), vascular endothelial growth factor receptor (VEGFR), human epidermal growth factor receptor-2 (HER2), and others, they failed to be effective therapeutic targets for ESCC [11,12]. Since the pathogenesis of ESCC is still unclear, it is necessary to identify novel biomarkers for early diagnosis, oncology therapy, and prognosis prediction.
Exocyst complex component 3-like 1 (EXOC3L1) is a protein-coding RNA located on chromosome 16 (16q22.1), also known as EXOC3L, which was originally isolated and reported as an isoform of Sec6 to regulate insulin secretion in pancreatic β cells. Previous studies suggested that EXOC3L1 is associated with insulin secretion, high-density lipoprotein (HDL) concentration [13], and spontaneously-induced apoptosis [14]. However, few studies on EXOC3L1 have been reported, EXOC3L1 has not been studied in any cancers so far, and the association between EXOC3L1 and ESCC has not yet been characterized, and these topics warrant further investigation. The purpose of the present study was to investigate the relationship between EXOC3L1 and ESCC.
Material and Methods
DATA SOURCE:
The RNA-seq data were downloaded from the University of California Santa Cruz (UCSC) Xena (
DIFFERENTIALLY EXPRESSED GENES (DEGS) ANALYSIS:
EXOC3L1-related DEGs were identified in 82 ESCC samples by the DESeq2 R package [15] using Wilcoxon rank-sum test, and the cut-off value was 50%. The DEGs met the requirements of |logFC| ≥1.5, and P values <0.05 were considered as significantly different DEGs. The relationship between EXOC3L1 and the top 10 upregulated genes were displayed by a heatmap, and the significantly different DEGs were used for subsequent enrichment analysis and protein–protein interaction (PPI) network analysis.
FUNCTIONAL ENRICHMENT ANALYSIS:
In this study, the significantly different DEGs were chosen to further perform functional enrichment analysis by using clusterProfiler R package [16], which included Gene Ontology (GO) functional analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, and gene set enrichment analysis (GSEA).
PPI NETWORK ANALYSIS:
The PPI network of EXOC3L1-related DEGs was constructed using the search tool for the retrieval of interacting genes (STRING) online database (https://cn.string-db.org/) with an interaction threshold of 0.4, the interaction network was displayed by Cytoscape software (version 3.8.2) [17], and the most significant module in the PPI network was extracted by the MCODE plug-in.
IMMUNE INFILTRATION ANALYSIS:
The immune infiltration analysis of EXOC3L1 in ESCC was performed using the GSVA R package [18] using the single-sample GSEA (ssGSEA) method, the infiltration abundance of 24 types immune cells previously described [19] were quantified, and the immune infiltration difference between the high EXOC3L1 expression and low EXOC3L1 expression groups was taken into consideration as well.
STATISTICAL ANALYSIS:
All statistical analyses were performed using R software (v.3.6.3). The Wilcoxon rank-sum test was used to analyze the expression of EXOC3L1. Survival analysis was performed with the Kaplan-Meier method, and the difference between survival curves was compared with a log-rank test. Univariate and multivariate Cox regression analyses were performed to estimate the factors that contributed to the prognosis of ESCC. The Spearman test was applied to analyze the correlation between EXOC3L1 expression and immune cell infiltration. All statistical tests were 2-tailed and
Results
ABNORMALLY HIGH EXPRESSION OF EXOC3L1 IN ESCC:
The expression difference of EXOC3L1 was assessed in 652 normal samples from GTEx and 82 ESCC samples from TCGA. The results showed that EXOC3L1 was highly expressed in ESCC (Figure 1A). A receiver operating characteristic (ROC) curve was used to evaluate the discriminative power of EXOC3L1 expression to distinguish tumor tissues from normal tissues, with area under curve (AUC) of EXOC3L1 0.812 (Figure 1B), suggesting that EXOC3L1 could serve as an ideal biomarker for the diagnosis of ESCC. In addition, the expression of EXOC3L1 was assessed in the GEO database, showing that EXOC3L1 was significantly overexpressed in tumor tissues in the GSE53625 dataset (Figure 1C) and GSE161533 dataset (Figure 1D).
EXOC3L1 EXPRESSION WAS CORRELATED WITH THE PROGNOSIS OF ESCC:
The associations between EXOC3L1 expression and clinicopathological characteristics were investigated (Table 1). The results demonstrated that the expression of EXOC3L1 was correlated with the N stage (Figure 2A) and the pathologic stage (Figure 2B). To explore whether the expression of EXOC3L1 has an effect on the prognosis of the patients with ESCC, the associations between EXOC3L1 expression and OS, disease-specific survival (DSS), and progression-free interval (PFI) were evaluated by Kaplan-Meier method with log-rank test, showing that ESCC patients with high expression of EXOC3L1 had a worse prognosis than patients with low expression (Figure 2C). Next, univariate and multivariate Cox proportional hazards regression analyses were used to evaluate the factors influencing the OS, showing that pathologic stage and EXOC3L1 expression were independent predictive factors for OS in ESCC (Table 2).
IDENTIFICATION OF DEGS IN ESCC:
The DEGs between EXOC3L1 high-expression and low-expression groups were identified by the DSEeq2 R package in the RNA-sequencing data of ESCC in TCGA database (Figure 3A). The DEGs met the requirements of |logFC| ≥1.5, and P values <0.05 were considered as significantly different DEGs; 556 genes were obtained after filtering, covering 290 upregulated and 266 downregulated genes. The relationship between EXOC3L1 and the top 10 upregulated genes were displayed by a heatmap (Figure 3B), and the significantly different DEGs were used in functional enrichment analysis and then in the PPI network.
FUNCTIONAL ENRICHMENT ANALYSIS AND PPI NETWORK ANALYSIS BASED ON DEGS:
To explore the functions of the statistically significant DEGs in ESCC, conducted GO and KEGG functional enrichment analyses was performed using the clusterProfiler R package. GO analysis included biological process (BP), cellular composition (CC), and molecular function (MF). The results indicated that BP might be associated with “humoral immune response”, CC might be associated with “immunoglobulin complex”, and MF might be associated with “immunoglobulin receptor binding” (Figure 3C). KEGG pathway analysis showed that the “cytokine-cytokine receptor interaction” pathway might be involved in the biological process of ESCC (Figure 3D).
To obtain the interactions of the statistically significant DEGs, a PPI network was constructed using the STRING online database, and the interaction threshold was 0.40. Subsequently, the interaction was displayed with Cytoscape software (Figure 4A), and the most significant module in the PPI network was extracted by the MCODE plug-in (Figure 4B).
Additionally, GSEA was also performed to reveal the EXOC3L1-related pathways in ESCC, and the pathways met the requirements of P value <0.05, FDR <0.25, and |NES| ≥1 and were considered as significant different pathways. Among the significant different pathways, several were selected to exhibit, including intestinal immune network, T cell receptor signaling, Natural Killer T cell pathway, and cancer immunotherapy by PD1 blockade (Figure 5A–5D).
CORRELATION BETWEEN EXOC3L1 EXPRESSION AND IMMUNE INFILTRATION:
Finally, the correlations between the expression of EXOC3L1 and immune cell infiltrations were assessed by Spearman correlation test using ssGSEA method (Figure 6A). The findings revealed that the expression of EXOC3L1 was positively correlated with the abundance of active dendritic cell (aDC), CD8+ T cells, cytotoxic cells, eosinophils, immature dendritic cell (iDC), CD56dim Natural killer cells, T cells, Th1 cells, and regulatory T cells (Figure 6B, 6C).
Discussion
ESCC is one of the most aggressive cancers all over the world and the incidence continues to rise, despite the advances in treatment technology, the prognosis is still gloomy, and the treatment of ESCC is a worldwide challenge. Although numerous biomarkers have been investigated in ESCC, their reliability remains controversial, thus, it is essential to discover new biomarkers to perform early diagnosis, oncology therapy, and prognosis prediction of ESCC. EXOC3L1 has not yet been investigated in ESCC.
ESCC is rich in lymphatic vessels and is very susceptible to lymph node metastasis [20], which is the main mode of metastasis in ESCC, and more than half of patients already have multiple lymph node metastasis at the time of initial diagnosis [21]. The presence of lymph node metastasis means a higher stage and even loss of the opportunity for radical surgery, with a gloomy prognosis [22]. Therefore, understanding the mechanism of ESCC lymph node metastasis is a vital step in research on ESCC. The present study is the first to show that EXOC3L1 expression is correlated with the N stage in ESCC, suggesting that EXOC3L1 is closely related to lymph node metastasis and plays a crucial role in the mechanism of lymph node metastasis in ESCC. This important discovery helps to clarify the mechanism of lymph node metastasis in ESCC, and may provide a new target for the treatment of ESCC.
In addition, the relationship between EXOC3L1 and ESCC prognosis has not been reported previously, and the present study demonstrated that EXOC3L1 was significantly elevated in ESCC samples, which indicated poorer clinical outcomes, and the OS, DSS, and PFI were significantly worse among patients with high EXOC3L1 expression than in those with low expression. Multivariate Cox regression analysis further demonstrated that EXOC3L1 could act as an independent prognostic indicator of ESCC. There have been few recent breakthroughs in targeted therapy of ESCC [7,8,10], and the lack of effective targeted drugs is one of the main reasons for the poor prognosis of ESCC [23]. The present study found that EXOC3L1 was significantly increased in ESCC and was significantly related to the prognosis of ESCC patients, which means that EXOC3L1 has the potential to be a novel target for ESCC. Further in-depth research on the pathogenesis of EXOC3L1 in ESCC is expected to provide new insights for the treatment of ESCC.
It is well known that the immune microenvironment plays a crucial role in the occurrence and development of tumors [24,25], the changes in the immune microenvironment may be the initiating factors for many pathophysiological, and the mechanism of immune microenvironment is extremely complicated [26,27]. In the present study, GO analysis, KEGG analysis, and GSEA analysis all suggested that EXOC3L1 is related to the immune function of ESCC, which was consistent with the immune infiltration analysis, showing that high expression of EXOC3L1 was positively correlated with an increase in immune infiltration levels in activated dendritic cells, CD8+ T cells, cytotoxic cells, eosinophils, immature dendritic cells, natural killer CD56dim cells, T cells, Th1 cells, and regulatory T cells. Although some previous studies reported that most immune cells have a positive impact on clinical outcomes [28–32], this is not totally consistent with the results of the present study. A reasonable explanation for the disagreement is that the changing immune microenvironment is very complex, immune cells in the tumor microenvironment have dual effects of anti-tumor and tumor-promoting [33,34], and immune cells can perform different functions in different tumors or at different stages of development in the same type of tumor. To date, clinical trials of targeted research in ESCC have not achieved ideal results, while the clinical trials of immunotherapy in ESCC show promise, and immunotherapy has shown great potential in the treatment of ESCC [35–38], suggesting that the therapeutic strategies targeting changes in the immune microenvironment are likely to benefit patients with ESCC; thus, the present study provides promising insights for the immunotherapy of ESCC by manipulating the immune microenvironment.
The highlights of the present study are as follows. Above all, EXOC3L1 has not been investigated in any tumors so far and this was the first study to reveal the relationship between EXOC3L1 and ESCC. The function of EXOC3L1 as an oncogene in ESCC was initially clarified, which could affect the metastasis and prognosis of ESCC patients and was related to immune infiltration, laying a foundation for further study of the mechanism of EXOC3L1 in ESCC in the future. Another important aspect of this study was that the expression of EXOC3L1 in ESCC was investigated in GTEx and TCGA, and was confirmed by GEO database and paired fresh tissues, which made the conclusion more reliable.
Although this study improves understanding of the relationship between EXOC3L1 and ESCC, there were still some unavoidable limitations. Firstly, the research was performed mainly based on bioinformatics analysis, so further verification should be carried out to reveal the biological impacts of EXOC3L1 in ESCC by in vitro and in vivo experiments. Secondly, a retrospective study has its own disadvantages, especially in that the interventions were not uniform and some of the information was missing; thus, a prospective study should be performed in the future to avoid analysis bias arising from the retrospective design. The third shortcoming of this study was that the sample size was small, and to enhance the credibility of the results, the sample size should be expanded.
Conclusions
In summary, the present study assessed the prognostic value of EXOC3L1 in ESCC for the first time, suggesting that EXOC3L1 offers a promising biomarker for predicting the prognosis and is a potential therapeutic target for ESCC patients. However, to elucidate the biological impact and underlying mechanism of EXOC3L1 in ESCC, further experimental studies are warranted.
Figures
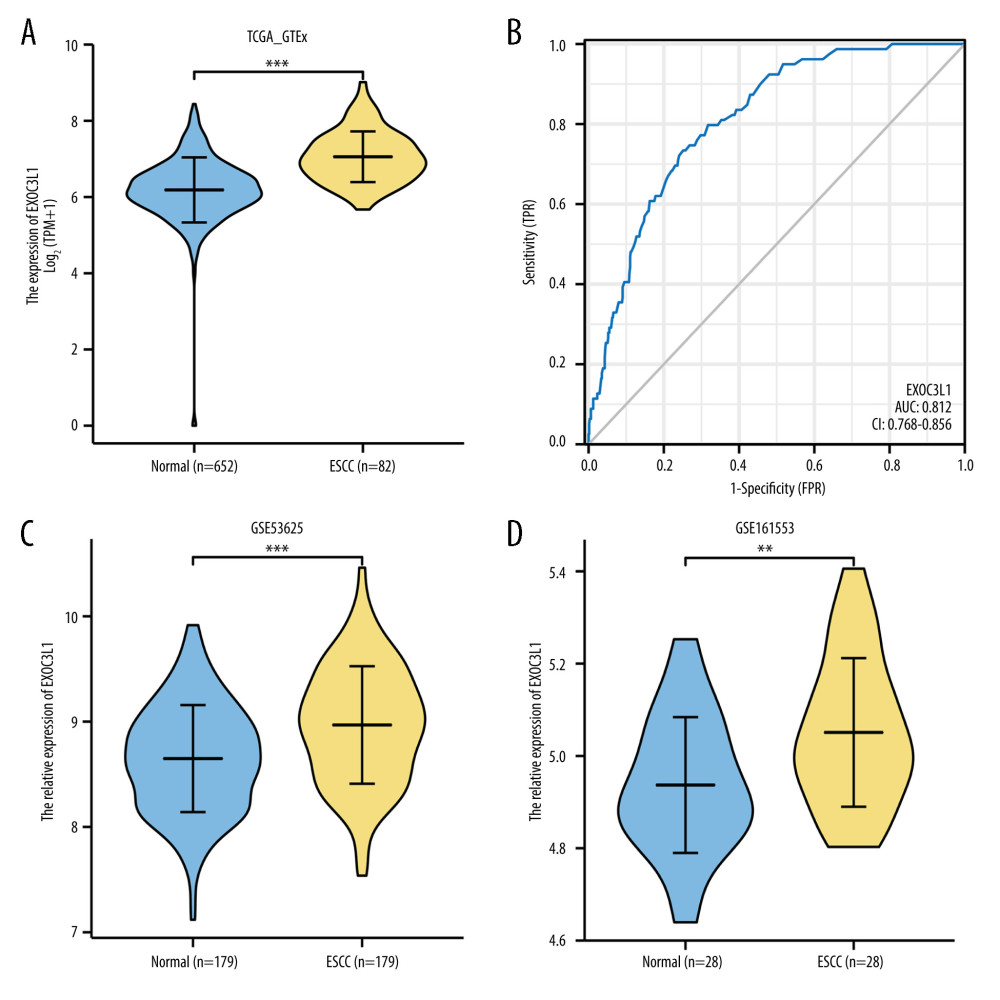 Figure 1. EXOC3L1 was highly expressed in ESCC(A) EXOC3L1 was significantly overexpressed in ESCC compared to normal samples. (B) The ROC curve of EXOC3L1 in the TCGA_GTEx dataset showed an AUC value of 0.812. (C) In the GSE53625 dataset, EXOC3L1 expression was higher in tumors than in normal tissues. (D) EXOC3L1 was upregulated in tumor tissues compared with normal tissues in the samples from GSE161533. * P<0.05, ** P<0.01, *** P<0.001.
Figure 1. EXOC3L1 was highly expressed in ESCC(A) EXOC3L1 was significantly overexpressed in ESCC compared to normal samples. (B) The ROC curve of EXOC3L1 in the TCGA_GTEx dataset showed an AUC value of 0.812. (C) In the GSE53625 dataset, EXOC3L1 expression was higher in tumors than in normal tissues. (D) EXOC3L1 was upregulated in tumor tissues compared with normal tissues in the samples from GSE161533. * P<0.05, ** P<0.01, *** P<0.001. 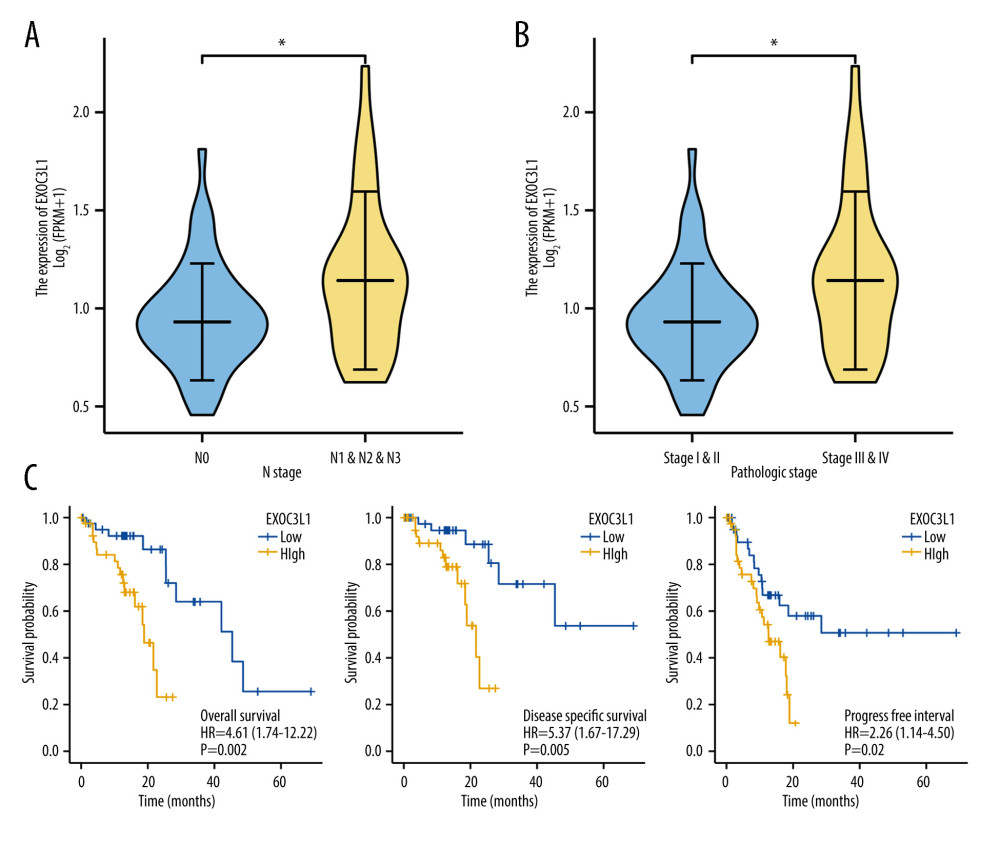 Figure 2. High expression of EXOC3L1 was associated with worse prognosis in patients with ESCC(A) Among the ESCC patients from TCGA, the expression of EXOC3L1 was significantly correlated with N stage. (B) High expression of EXOC3L1 was associated with higher pathological stage. (C) The ESCC patients with high EXOC3L1 expression had a shorter OS, DSS, and PFI.
Figure 2. High expression of EXOC3L1 was associated with worse prognosis in patients with ESCC(A) Among the ESCC patients from TCGA, the expression of EXOC3L1 was significantly correlated with N stage. (B) High expression of EXOC3L1 was associated with higher pathological stage. (C) The ESCC patients with high EXOC3L1 expression had a shorter OS, DSS, and PFI. 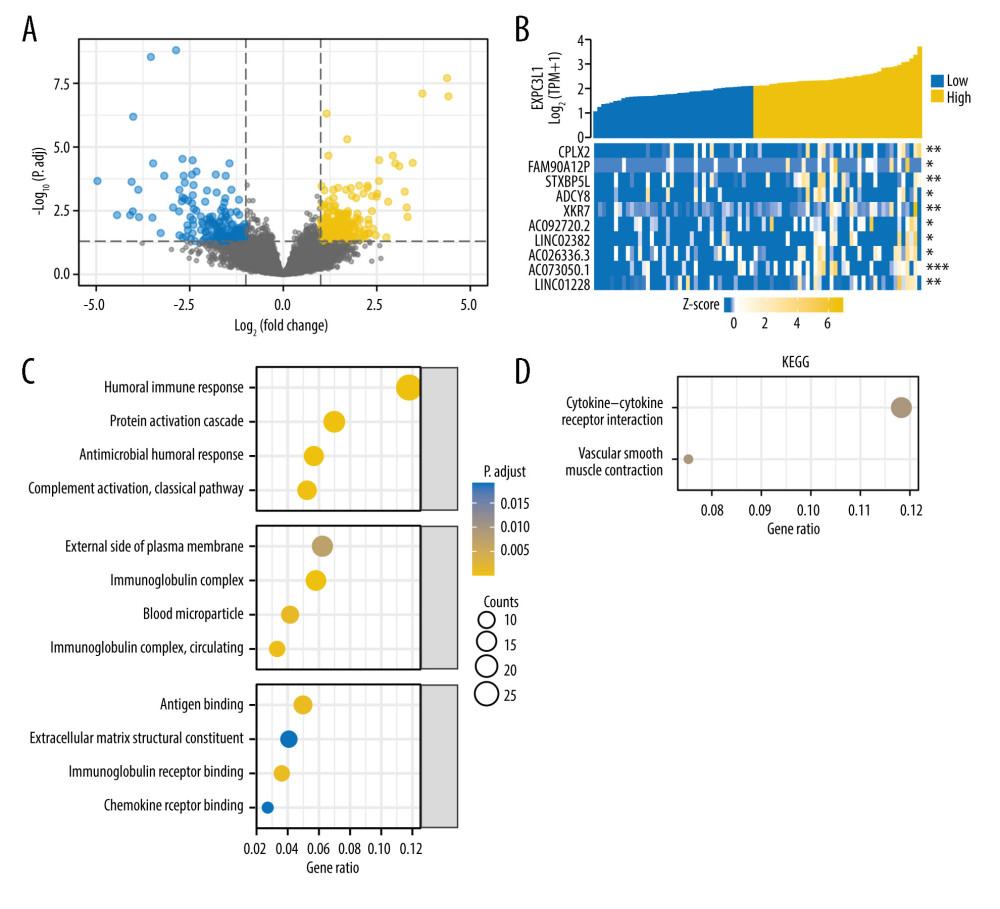 Figure 3. GO and KEGG analysis based on the DEGs(A) The differential expression analysis of EXOC3L1 in 82 ESCC samples from TCGA, blue representing downregulated expression and yellow representing upregulated expression. (B) The relationship between the top 10 upregulated genes and the EXOC3L1 expression in different groups was displayed by a heatmap. (C) GO enrichment analysis, including BP, CC, and MF. (D) KEGG pathway annotations.
Figure 3. GO and KEGG analysis based on the DEGs(A) The differential expression analysis of EXOC3L1 in 82 ESCC samples from TCGA, blue representing downregulated expression and yellow representing upregulated expression. (B) The relationship between the top 10 upregulated genes and the EXOC3L1 expression in different groups was displayed by a heatmap. (C) GO enrichment analysis, including BP, CC, and MF. (D) KEGG pathway annotations. 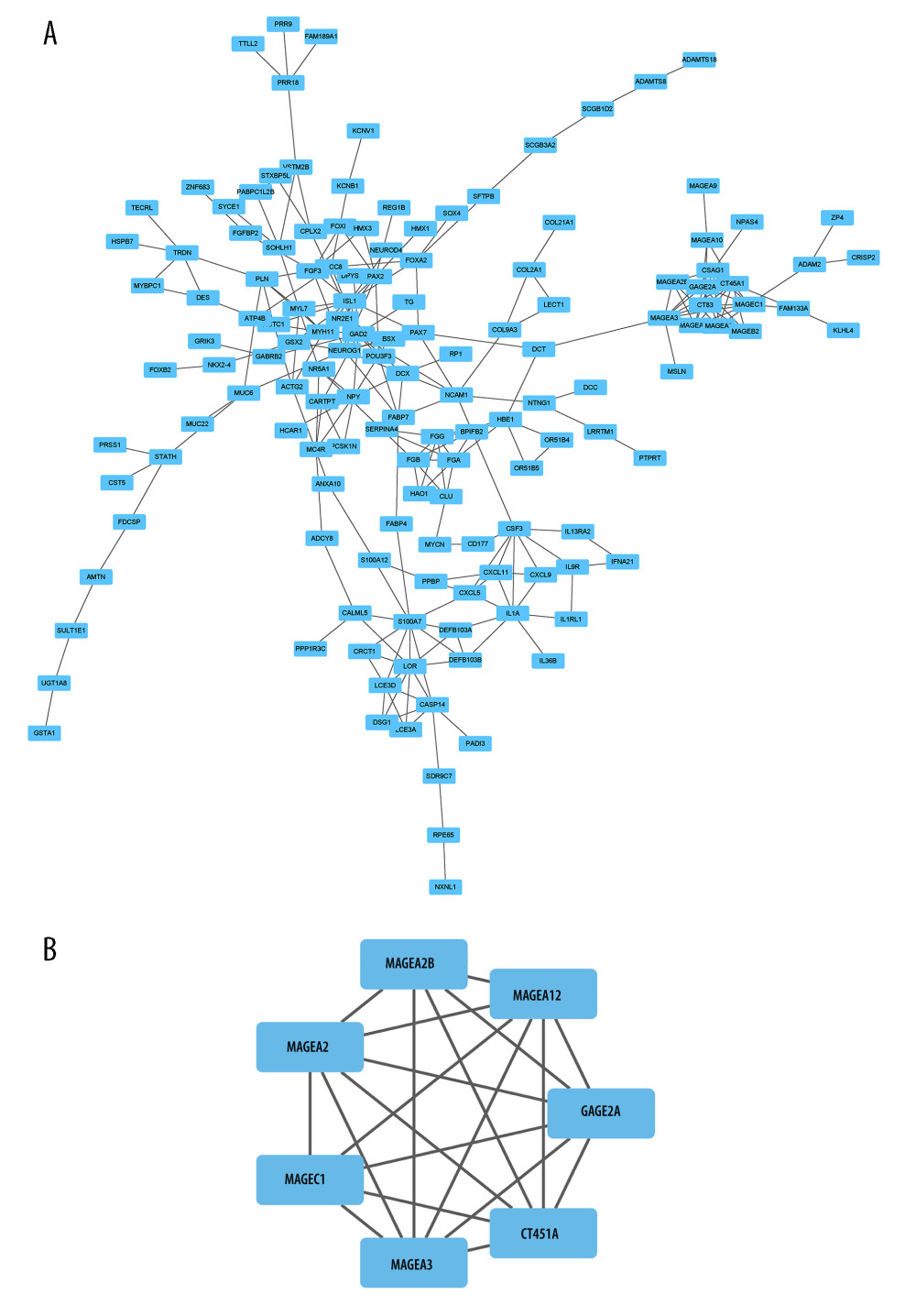 Figure 4. PPI network analysis based on the DEGs(A) PPI network analysis of DEGs. (B) The most significant module in the PPI network was identified by the MCODE plug-in.
Figure 4. PPI network analysis based on the DEGs(A) PPI network analysis of DEGs. (B) The most significant module in the PPI network was identified by the MCODE plug-in. 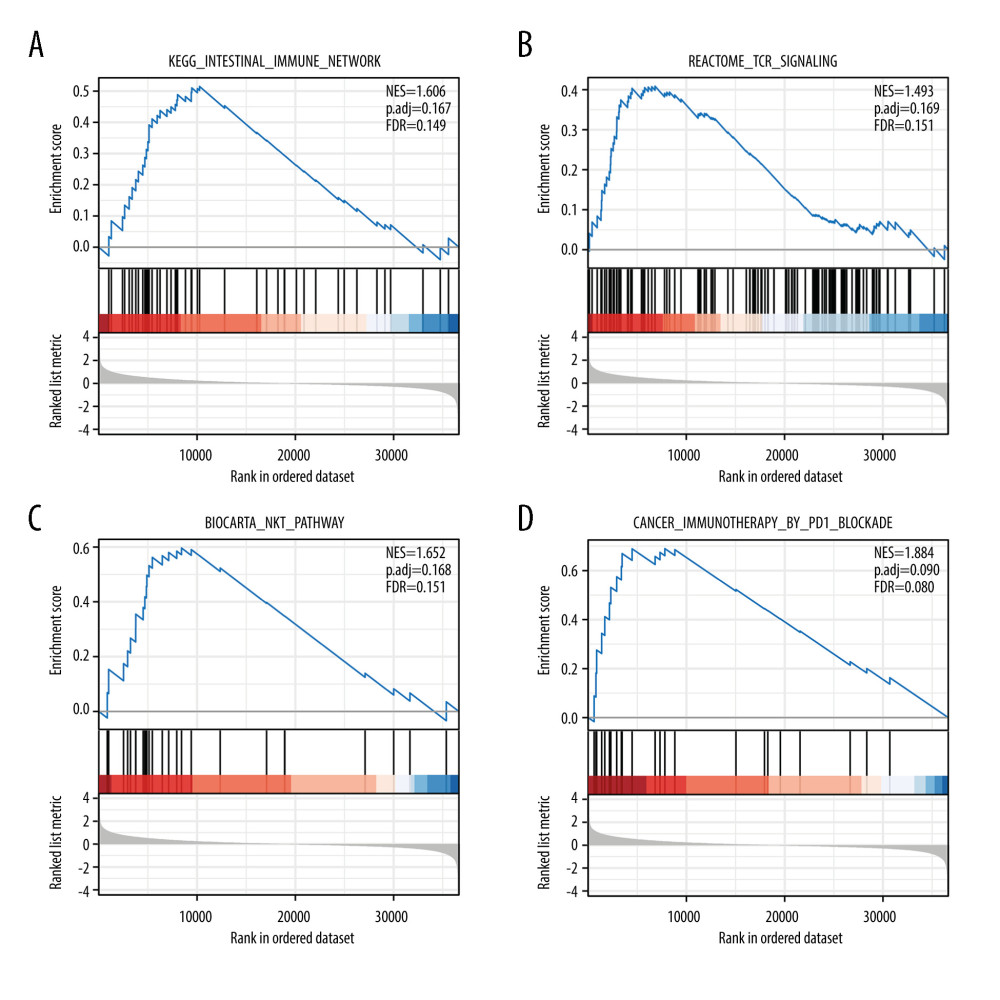 Figure 5. Enrichment plots of the GSEA(A) Intestinal immune network. (B) T cell receptor signaling. (C) Natural killer T cell pathway. (D) Cancer immunotherapy by PD1 blockade.
Figure 5. Enrichment plots of the GSEA(A) Intestinal immune network. (B) T cell receptor signaling. (C) Natural killer T cell pathway. (D) Cancer immunotherapy by PD1 blockade. 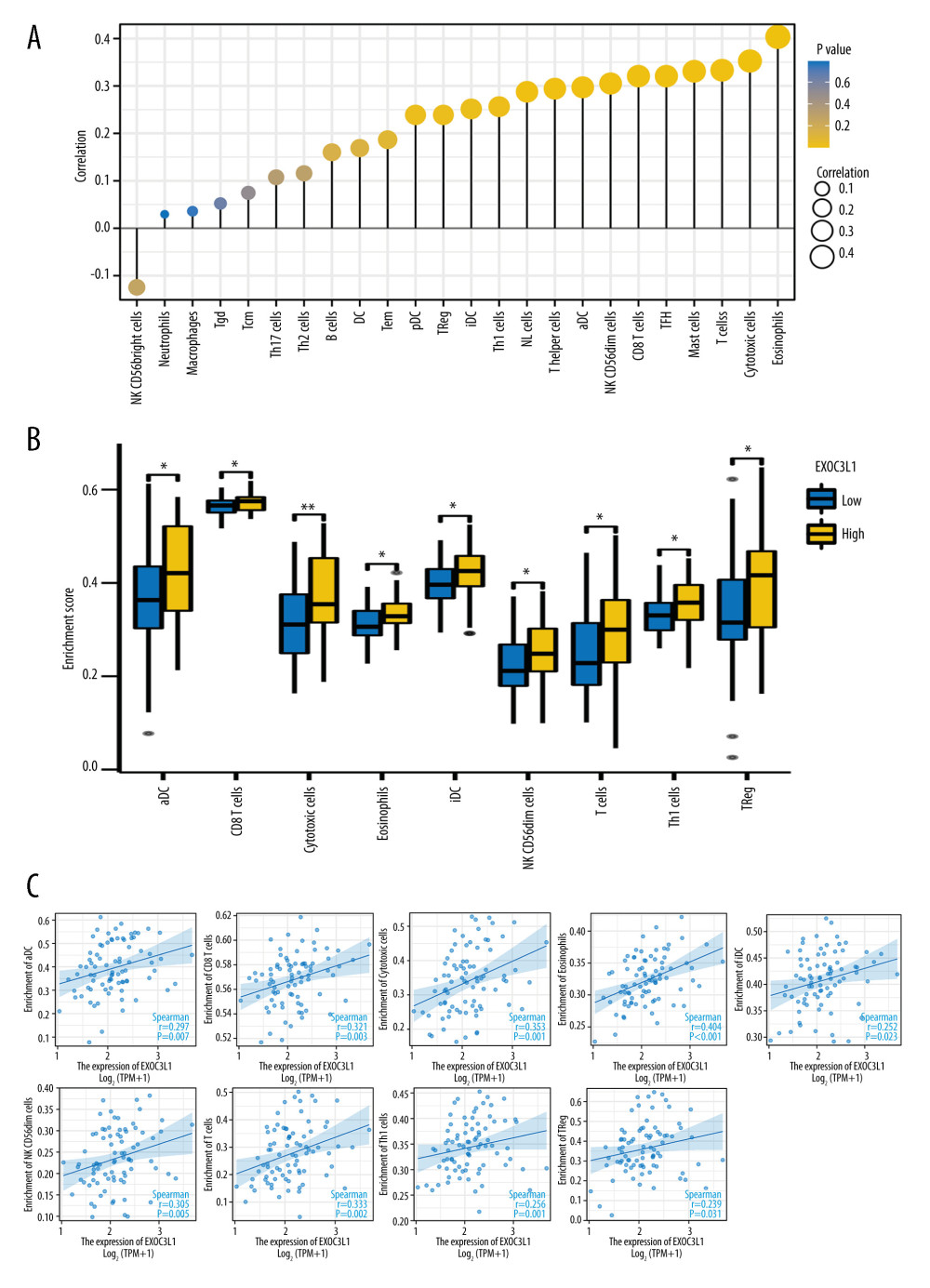 Figure 6. The expression of EXOC3L1 was associated with immune infiltration in ESCC(A) The correlation between EXOC3L1 expression and 24 kinds of immune cells. (B, C) The expression level of EXOC3L1 was significantly correlated with the abundance of several types of immune cells (B), including activated dendritic cells, CD8+ T cells, cytotoxic cells, eosinophils, immature dendritic cells, natural killer CD56dim cells, T cells, Th1 cells, and regulatory T cells (C). * P<0.05, ** P<0.01, *** P<0.001.
Figure 6. The expression of EXOC3L1 was associated with immune infiltration in ESCC(A) The correlation between EXOC3L1 expression and 24 kinds of immune cells. (B, C) The expression level of EXOC3L1 was significantly correlated with the abundance of several types of immune cells (B), including activated dendritic cells, CD8+ T cells, cytotoxic cells, eosinophils, immature dendritic cells, natural killer CD56dim cells, T cells, Th1 cells, and regulatory T cells (C). * P<0.05, ** P<0.01, *** P<0.001. References
1. Bray F, Ferlay J, Soerjomataram I, Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries: Cancer J Clin, 2018; 68(6); 394-424
2. Lagergren J, Smyth E, Cunningham D, Lagergren P, Oesophageal cancer: Lancet, 2017; 390(10110); 2383-96
3. Smyth EC, Lagergren J, Fitzgerald RC, Oesophageal cancer: Nat Rev Dis Primers, 2017; 3; 17048
4. Abnet CC, Arnold M, Wei WQ, Epidemiology of esophageal squamous cell carcinoma: Gastroenterology, 2018; 154(2); 360-73
5. DiSiena M, Perelman A, Birk J, Rezaizadeh H, Esophageal cancer: An updated review: South Med J, 2021; 114(3); 161-68
6. Kashyap MK, Abdel-Rahman O, Expression, regulation and targeting of receptor tyrosine kinases in esophageal squamous cell carcinoma: Mol Cancer, 2018; 17(1); 54
7. Harada K, Rogers JE, Iwatsuki M, Recent advances in treating oesophageal cancer: F1000Res, 2020; 9; F1000 F aculty Rev-1189
8. He S, Xu J, Liu X, Zhen Y, Advances and challenges in the treatment of esophageal cancer: Acta Pharm Sin B, 2021; 11(11); 3379-92
9. Fatehi Hassanabad A, Chehade R, Breadner D, Raphael J, Esophageal carcinoma: Towards targeted therapies: Cell Oncol (Dordr), 2020; 43(2); 195-209
10. Yang YM, Hong P, Xu WW, Advances in targeted therapy for esophageal cancer: Signal Transduct Target Ther, 2020; 5(1); 229
11. van Rossum PSN, Mohammad NH, Vleggaar FP, van Hillegersberg R, Treatment for unresectable or metastatic oesophageal cancer: current evidence and trends: Nat Rev Gastroenterol Hepatol, 2018; 15(4); 235-49
12. Hirano H, Kato K, Systemic treatment of advanced esophageal squamous cell carcinoma: Chemotherapy, molecular-targeting therapy and immunotherapy: Jpn J Clin Oncol, 2019; 49(5); 412-20
13. Lanktree MB, Elbers CC, Li Y, Genetic meta-analysis of 15,901 African Americans identifies variation in EXOC3L1 is associated with HDL concentration: J Lipid Res, 2015; 56(9); 1781-86
14. Matsumoto S, Okada J, Yamada E, Overexpressed exocyst complex component 3-like 1 spontaneously induces apoptosis: Biomed Res, 2021; 42(3); 109-13
15. Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2: Genome Biol, 2014; 15(12); 550
16. Yu G, Wang LG, Han Y, He QY, clusterProfiler: An R package for comparing biological themes among gene clusters: OMICS, 2012; 16(5); 284-87
17. Otasek D, Morris JH, Boucas J, Cytoscape automation: Empowering workflow-based network analysis: Genome Biol, 2019; 20(1); 185
18. Hnzelmann S, Castelo R, Guinney JJBB, GSVA: Gene set variation analysis for microarray and RNA-Seq data: BMC Bioinformatics, 2013; 14(1); 7
19. Bindea G, Mlecnik B, Tosolini M, Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer: Immunity, 2013; 39(4); 782-95
20. Li J, Qi Z, Hu YP, Wang YX, Possible biomarkers for predicting lymph node metastasis of esophageal squamous cell carcinoma: A review: J Int Med Res, 2019; 47(2); 544-56
21. Weiss F, Lauffenburger D, Friedl P, Towards targeting of shared mechanisms of cancer metastasis and therapy resistance: Nat Rev Cancer, 2022; 22(3); 157-73
22. Ohashi S, Miyamoto S, Kikuchi O, Recent advances from basic and clinical studies of esophageal squamous cell carcinoma: Gastroenterology, 2015; 149(7); 1700-15
23. Lin DC, Wang MR, Koeffler HP, Genomic and epigenomic aberrations in esophageal squamous cell carcinoma and implications for patients: Gastroenterology, 2018; 154(2); 374-89
24. Zheng Y, Chen Z, Han Y, Immune suppressive landscape in the human esophageal squamous cell carcinoma microenvironment: Nat Commun, 2020; 11(1); 6268
25. Lian J, Yue Y, Yu W, Zhang Y, Immunosenescence: A key player in cancer development: J Hematol Oncol, 2020; 13(1); 151
26. Deng YJ, Ren EH, Yuan WH, GRB10 and E2F3 as diagnostic markers of osteoarthritis and their correlation with immune infiltration: Diagnostics (Basel), 2020; 10(3); 171
27. Dieci MV, Miglietta F, Guarneri V, Immune infiltrates in breast cancer: Recent updates and clinical implications: Cells, 2021; 10(2); 223
28. Tosolini M, Kirilovsky A, Mlecnik B, Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer: Cancer Res, 2011; 71(4); 1263-71
29. Gajewski TF, Schreiber H, Fu YX, Innate and adaptive immune cells in the tumor microenvironment: Nat Immunol, 2013; 14(10); 1014-22
30. Mu L, Ding K, Tu R, Yang W, Identification of 4 immune cells and a 5-lncRNA risk signature with prognosis for early-stage lung adenocarcinoma: J Transl Med, 2021; 19(1); 127
31. Grisaru-Tal S, Itan M, Klion AD, Munitz A, A new dawn for eosinophils in the tumour microenvironment: Nat Rev Cancer, 2020; 20(10); 594-607
32. Fu C, Jiang A, Dendritic cells and CD8 T cell immunity in tumor microenvironment: Front Immunol, 2018; 9; 3059
33. Pena-Romero AC, Orenes-Pinero E, Dual effect of immune cells within tumour microenvironment: pro- and anti-tumour effects and their triggers: Cancers (Basel), 2022; 14(7); 1681
34. Dou A, Fang J, Heterogeneous myeloid cells in tumors: Cancers (Basel), 2021; 13(15); 3772
35. Baba Y, Nomoto D, Okadome K, Tumor immune microenvironment and immune checkpoint inhibitors in esophageal squamous cell carcinoma: Cancer Sci, 2020; 111(9); 3132-41
36. Huang TX, Fu L, The immune landscape of esophageal cancer: Cancer Commun (Lond), 2019; 39(1); 79
37. Doki Y, Ajani JA, Kato K, Nivolumab combination therapy in advanced esophageal squamous-cell carcinoma: N Engl J Med, 2022; 386(5); 449-62
38. Kelly RJ, Ajani JA, Kuzdzal J, Adjuvant nivolumab in resected esophageal or gastroesophageal junction cancer: N Engl J Med, 2021; 384(13); 1191-203
Figures
 Figure 1. EXOC3L1 was highly expressed in ESCC(A) EXOC3L1 was significantly overexpressed in ESCC compared to normal samples. (B) The ROC curve of EXOC3L1 in the TCGA_GTEx dataset showed an AUC value of 0.812. (C) In the GSE53625 dataset, EXOC3L1 expression was higher in tumors than in normal tissues. (D) EXOC3L1 was upregulated in tumor tissues compared with normal tissues in the samples from GSE161533. * P<0.05, ** P<0.01, *** P<0.001.
Figure 1. EXOC3L1 was highly expressed in ESCC(A) EXOC3L1 was significantly overexpressed in ESCC compared to normal samples. (B) The ROC curve of EXOC3L1 in the TCGA_GTEx dataset showed an AUC value of 0.812. (C) In the GSE53625 dataset, EXOC3L1 expression was higher in tumors than in normal tissues. (D) EXOC3L1 was upregulated in tumor tissues compared with normal tissues in the samples from GSE161533. * P<0.05, ** P<0.01, *** P<0.001. Figure 2. High expression of EXOC3L1 was associated with worse prognosis in patients with ESCC(A) Among the ESCC patients from TCGA, the expression of EXOC3L1 was significantly correlated with N stage. (B) High expression of EXOC3L1 was associated with higher pathological stage. (C) The ESCC patients with high EXOC3L1 expression had a shorter OS, DSS, and PFI.
Figure 2. High expression of EXOC3L1 was associated with worse prognosis in patients with ESCC(A) Among the ESCC patients from TCGA, the expression of EXOC3L1 was significantly correlated with N stage. (B) High expression of EXOC3L1 was associated with higher pathological stage. (C) The ESCC patients with high EXOC3L1 expression had a shorter OS, DSS, and PFI. Figure 3. GO and KEGG analysis based on the DEGs(A) The differential expression analysis of EXOC3L1 in 82 ESCC samples from TCGA, blue representing downregulated expression and yellow representing upregulated expression. (B) The relationship between the top 10 upregulated genes and the EXOC3L1 expression in different groups was displayed by a heatmap. (C) GO enrichment analysis, including BP, CC, and MF. (D) KEGG pathway annotations.
Figure 3. GO and KEGG analysis based on the DEGs(A) The differential expression analysis of EXOC3L1 in 82 ESCC samples from TCGA, blue representing downregulated expression and yellow representing upregulated expression. (B) The relationship between the top 10 upregulated genes and the EXOC3L1 expression in different groups was displayed by a heatmap. (C) GO enrichment analysis, including BP, CC, and MF. (D) KEGG pathway annotations. Figure 4. PPI network analysis based on the DEGs(A) PPI network analysis of DEGs. (B) The most significant module in the PPI network was identified by the MCODE plug-in.
Figure 4. PPI network analysis based on the DEGs(A) PPI network analysis of DEGs. (B) The most significant module in the PPI network was identified by the MCODE plug-in. Figure 5. Enrichment plots of the GSEA(A) Intestinal immune network. (B) T cell receptor signaling. (C) Natural killer T cell pathway. (D) Cancer immunotherapy by PD1 blockade.
Figure 5. Enrichment plots of the GSEA(A) Intestinal immune network. (B) T cell receptor signaling. (C) Natural killer T cell pathway. (D) Cancer immunotherapy by PD1 blockade. Figure 6. The expression of EXOC3L1 was associated with immune infiltration in ESCC(A) The correlation between EXOC3L1 expression and 24 kinds of immune cells. (B, C) The expression level of EXOC3L1 was significantly correlated with the abundance of several types of immune cells (B), including activated dendritic cells, CD8+ T cells, cytotoxic cells, eosinophils, immature dendritic cells, natural killer CD56dim cells, T cells, Th1 cells, and regulatory T cells (C). * P<0.05, ** P<0.01, *** P<0.001.
Figure 6. The expression of EXOC3L1 was associated with immune infiltration in ESCC(A) The correlation between EXOC3L1 expression and 24 kinds of immune cells. (B, C) The expression level of EXOC3L1 was significantly correlated with the abundance of several types of immune cells (B), including activated dendritic cells, CD8+ T cells, cytotoxic cells, eosinophils, immature dendritic cells, natural killer CD56dim cells, T cells, Th1 cells, and regulatory T cells (C). * P<0.05, ** P<0.01, *** P<0.001. Tables
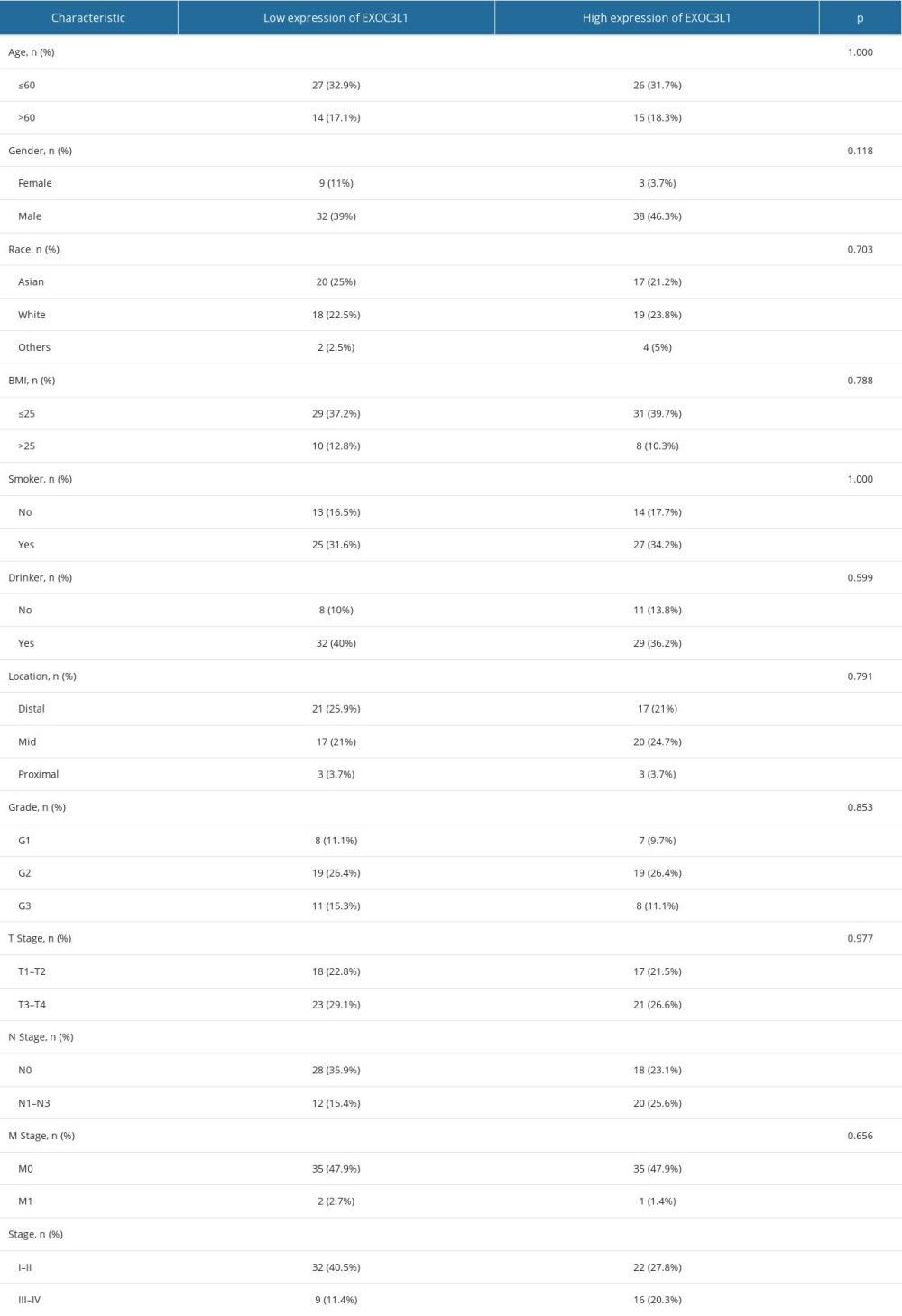 Table 1. The correlation between the expression of EXOC3L1 and clinical parameters in ESCC.
Table 1. The correlation between the expression of EXOC3L1 and clinical parameters in ESCC.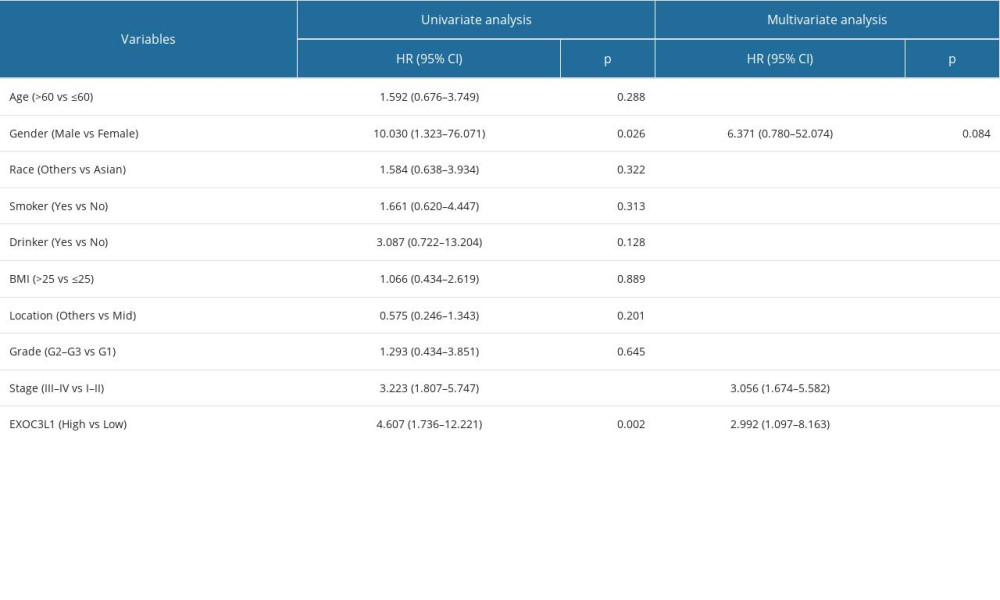 Table 2. Univariate and multivariate Cox regression analysis of overall survival in patients with ESCC.
Table 2. Univariate and multivariate Cox regression analysis of overall survival in patients with ESCC. Table 1. The correlation between the expression of EXOC3L1 and clinical parameters in ESCC.
Table 1. The correlation between the expression of EXOC3L1 and clinical parameters in ESCC. Table 2. Univariate and multivariate Cox regression analysis of overall survival in patients with ESCC.
Table 2. Univariate and multivariate Cox regression analysis of overall survival in patients with ESCC. In Press
05 Mar 2024 : Clinical Research
Muscular Function Recovery from General Anesthesia in 132 Patients Undergoing Surgery with Acceleromyograph...Med Sci Monit In Press; DOI: 10.12659/MSM.942780
05 Mar 2024 : Clinical Research
Effects of Thermal Insulation on Recovery and Comfort of Patients Undergoing Holmium Laser LithotripsyMed Sci Monit In Press; DOI: 10.12659/MSM.942836
05 Mar 2024 : Clinical Research
Role of Critical Shoulder Angle in Degenerative Type Rotator Cuff Tears: A Turkish Cohort StudyMed Sci Monit In Press; DOI: 10.12659/MSM.943703
06 Mar 2024 : Clinical Research
Comparison of Outcomes between Single-Level and Double-Level Corpectomy in Thoracolumbar Reconstruction: A ...Med Sci Monit In Press; DOI: 10.12659/MSM.943797
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952








