09 September 2023: Clinical Research
Synergistic Role of Biofilm-Associated Genes and Efflux Pump Genes in Tigecycline Resistance of
Bin Luo1AE*, Zhiwei Li1B, Qian Wang2C, Changmin Wang2FDOI: 10.12659/MSM.940704
Med Sci Monit 2023; 29:e940704
Abstract
BACKGROUND: Previous research reported that the resistance mechanism of Acinetobacter baumannii resistance to tigecycline was mainly related to the overexpression of the AdeABC efflux pump system. Biofilm formation is a notable pathogenesis of A. baumannii infections and antibiotic resistance. Our study explores the latent relevance of biofilm-associated genes and efflux pump genes in A. baumannii tigecycline resistance.
MATERIAL AND METHODS: A total of 78 clinical samples were collected from October 2018 to October 2019. Seventy-two clinically isolated A. baumannii strains were divided into a tigecycline-resistant Acinetobacter baumannii (TR-AN) group and tigecycline-sensitive Acinetobacter baumannii (TS-AN) group by tigecycline minimum inhibitory concentration tests. The biofilm formation of the 2 groups was observed using crystal violet staining. Furthermore, biofilm-related genes and efflux pump genes were analyzed by RT-PCR.
RESULTS: The biofilm-forming rate of the TR-AN group was 82.2%, and that of the TS-AN group was 14.8%. The biofilm synthesis gene bfs was 91.3% positive in the TR-AN group, significantly higher than in the TS-AN group at the transcription level (P<0.05). The minimum inhibitory concentration of tigecycline was higher in the TR-AN group with biofilm formation than in the TR-AN group without biofilm formation (P<0.05). The efflux pump AdeB gene was 95.2% positive in the TR-AN group with biofilm formation and 38.7% positive in the TR-AN group without biofilm formation.
CONCLUSIONS: The biofilm formation of A. baumannii may be positively related to tigecycline resistance ability because of the co-expression of the bfs gene and the AdeB efflux pump gene. The enhanced transcription level of bfs and AdeB promotes biofilm formation to improve the resistance of A. baumannii to tigecycline.
Keywords: AdeC Protein, Acinetobacter baumannii, AdeB Protein, Acinetobacter baumannii, AdeA Protein, Acinetobacter baumannii, Acinetobacter baumannii, Tigecycline, Biofilms, Cell Aggregation, Gentian Violet
Background
At present, it is mainly considered that the increased expression of the RND efflux pump gene is one of the important mechanisms of tigecycline resistance in
Greene et al [7] found that environmental isolates produced more biofilm than did clinical patient isolates. In clinical isolates, the multi-drug resistance phenotype increases their resistance to the environment, and the high biofilm phenotype synergizes to improve tolerance. In environmental isolates, the multi-drug resistance phenotype carries a fitness cost of reducing desiccation tolerance, which is buffered by the high biofilm phenotype. The expression of biofilm genes can vary according to environmental conditions, suggesting that the potential of
Tigecycline is a new class of tetracycline anti-infective drugs for intravenous injection, developed by Wyeth Pharmaceuticals and derived from minocycline [8,9]. Tigecycline is the first ammonia acyl antibiotic approved by the FDA for clinical use for complex abdominal cavity infection, skin infection, and the treatment of community-acquired pneumonia [10,11]. Tigecycline has an excellent effect on
In this study, we investigated 78 clinical samples at the Clinical Center of People’s Hospital of Xinjiang Uygur Autonomous Region from October 2018 to October 2019 and isolated 72
Material and Methods
ETHICAL APPROVAL:
All procedures in this study were conducted and approved by the Ethics Committee of the People’s Hospital of Xinjiang Uygur Autonomous Region. Written informed consent was obtained from the patients for their anonymized information to be published in this article.
BACTERIAL STRAINS AND CLINICAL SAMPLES:
Seventy-two A. baumannii strains were collected from clinical specimens between October 2018 and October 2019 at the Clinical Laboratory Department of Xinjiang Regional Hospital, as identified by the VITEK Rapid Bacterial Identification card and VITEK Mass Spestrometry MALD-TOF (bioMérieux Clinical Diagnostics, France) [19,20], according to the Database from the American Society for Microbiology. The specimens then grew on standard MacConkey agar (Neogen, USA) at 37°C for 48 h. The strains were obtained from blood, 21 (29.2%); urine, 13 (18.1%); and 38 (52.8%) sputum. For all selected strains, duplicate strains of the same patient, samples of unknown type, and samples mixed with other Acinetobacter genera were excluded.
ANTIMICROBIAL SUSCEPTIBILITY TESTING:
The sensitivity of tigecycline was determined using VITEK2 Compat (bioMerieux Inc., France) by standard operating procedures, referring to recommended standards of the US FDA and Clinical Laboratory Standards Institute (CLSI), and was interpreted by the CLSI M100 2021 guidelines [21,22]. The MIC of tigecycline was detected by the broth microdilution method. When the MIC is less than 2 mg/L, it is considered sensitive; between 2 and 8 mg/L is considered intermediary; greater than 8 mg/L is considered drug resistant [23,24].
BIOFILM FORMATION ABILITY TESTING:
GENOME DNA EXTRACTION:
The genomic DNA of the
Genomic DNA preparations of
The tigecycline-sensitive and tigecycline-resistant groups were detected, the differences were compared, and the expression was detected by QT-PCR. An amount of 1000 mL 0.5× TAE buffer was prepared. The glue making tank and comb were prepared with buffer and rinsed with distilled water. An amount of 1.0 g agarose powder was weighed and poured into a clean conical flask; 100 mL 0.5× TAE buffer was added, and the mixture was heated in a microwave oven for 3 min to fully melt it into a uniform and transparent shape. The conical flask was rinsed with running water to quickly cool it to about 60°C, and 3 μL DNA green nucleic acid dye was added and mixed well. The comb was inserted into the glue making tank, while slowly pouring the agarose gel into the glue making mold so no bubbles were generated (bubbles removed with pipette). The gel was placed in the refrigerator at 4°C for 40 min to completely solidify it. The rubber block was put into the refrigerator with 0.5× TAE buffer in the electrophoresis tank, with attention paid to not generate bubbles at the sampling hole. An amount of 5 μL DNA marker was added in the first sampling well, and the PCR amplification products were added into each well in sequence for 5 μL. The positive and negative electrodes of the electrophoresis tank were correctly connected, and electrophoresis at 90 V constant voltage was conducted for 40 min. After electrophoresis, the power was turned off, the results in the gel imager were observed, and photos were taken.
BIOFILM-RELATED GENE AND EFFLUX PUMP GENES DETECTION:
The 8 biofilm synthesis-related genes and efflux pump genes were assessed by using the conventional PCR method. The primers of the target genes are listed in Table 1. The PCR products were further electrophoresed on a 1.5% (w/V) agarose gel, and grayscale was valued by Image J software.
MOLECULAR EVOLUTIONARY TREE ANALYSIS:
Gene amplification was performed by enterobacterial recombinant sequencing PCR (Enterobacterial repetitive intergenic consensus PCR [ERIC]) using the ERIC®-2 single primer sequence 5′-AAGTAAGTGACTGGGGTGAGCG-3′ [25]. Twenty-three A. baumannii colonies with genotypes of bfs were selected for homologous analysis using bioinformatics software. According to the Tenover criteria [26], the patterns were the same. For different subtypes with the same band, a difference of 2 to 3 bands is considered closely related, differences of 4 to 6 bands are considered possible correlations, and difference of more than 7 strips is considered not related.
STATISTICAL ANALYSIS:
GraphPad Prism9.0.0 software was used for statistical data processing. Average distribution data is represented by the mean and standard deviation (mean±SD). The comparison of biofilm formation ability of different groups showed a skewed distribution, and the results are expressed by the median and quartile range [M (P25, P75)]. A nonparametric rank-sum test was used to analyze the biological characteristics of strains. Biofilm formation ability and tigecycline resistance were analyzed using the chi-square test. All tests were 2-tailed. Medcal15.2 software was used to draw the receiver operating characteristic (ROC) curve and get the best cut-off value.
Results
BASELINE CHARACTERISTICS:
As shown in Table 2, 45 men and 33 women were enrolled in the study. The patients’ average age was 60±21 years. Seventy-two A. baumannii strains were isolated. Among the 72 clinically isolated and cultured A. baumannii, 45 showed resistance to tigecycline. Additionally, 43.06% (31/72) of patients were admitted to the Intensive Care Unit, and 30.56% (22/72) had chronic obstructive pulmonary disease.
TIGECYCLINE SUSCEPTIBILITY:
The MICs of tigecycline were determined by the broth microdilution method. The tigecycline MIC of the 72 strains isolated ranged from 0.04 to 17.26 mg/L. There were 45 samples (62.5%) in the TR-AN (resistant) group and 27 (37.5%) in the TS-AN (sensitive) group (Table 3).
EFFLUX PUMP GENES ANALYSIS:
Transcription level analysis of blaOXA-23, blaOXA-58, blaOXA-51, AdeABC, and AdeRS genes showed AdeB expression in the TR-AN group was 6.1-fold higher than in the TS-AN group. AdeA, AdeC, and AdeRS genes increased 1.5, 2.1, and 0.7-fold, respectively, in the TR-AN group compared with in the TS-AN group (Figures 1, 2). AdeB gene expression of the TR-AN group was considerably higher than that of the TS-AN group (chi-square test, P<0.05).
Further transcriptional analysis of the AdeB gene in the TR-AN group with or without biofilm strains showed that the AbeB gene was 94.6% positive in the TR-AN group with biofilm formation and 37.5% positive in the TR-AN group without biofilm.
BIOFILM GROWTH DETECTION:
After 48 h of culture, the optical density value was measured and observed by laser confocal microscope; 37 strains of the 45 in the TR-AN group formed biofilms in the in vitro experiment, while in the TS-AN group, only 4 strains formed (Figure 3).
BIOFILM-RELATED GENE ANALYSIS:
In the 72 specimens, the detection rates of abaI, bfmS, bfmR, csuA, csuB, and csuE were all less than 20%, and those of bap, bfmR, and bfs were 34.7%, 41.6%, and 68.1%, respectively. A total of 37 strains of the 45 strains in the TR-AN group formed biofilms in the in vitro experiment, while in the TS-AN group, only 4 strains formed. By studying the transcriptional level of the bfs gene in the membrane-forming and the non-membrane-forming TR-AN groups, it can be concluded that the expression level of bfs in the biofilm-forming TR-AN group was 9.3-fold higher than that in the non-biofilm-forming TR-AN group. The results of biofilm-associated genes are presented in Table 4 and Figure 4.
GROWTH ABILITY OF BIOFILM WAS SIGNIFICANTLY DIFFERENT BETWEEN THE TR-AN AND TS-AN GROUPS:
Among the 72 clinically isolated A. baumannii strains, 57 formed biofilms in vitro. Regarding the ability of bacterial biofilm formation, the biofilm formation level of the TR-AN group was significantly higher than that of the TS-AN group (P<0.01; Table 5).
DIAGNOSTIC VALUE OF BIOFILM GROWTH ABILITY FOR TIGECYCLINE RESISTANCE:
The ROC curve was drawn, and the MIC value of tigecycline drug sensitivity was used as the standard group. The resistance of tigecycline was judged by the growth ability of biofilm. The upper and lower limits, group distance, and cut-off points of the measured values were determined by analyzing the ROC curves. The cumulative frequency distribution table was listed according to the selected group distance interval, and the sensitivity, specificity, and false positive rate (1-specificity) of all cut-off points were calculated respectively. The sensitivity as the ordinate represented the true positive rate, the 1-specificity as the abscissa represented the false positive rate, and the ROC curve was drawn.
The ROC curve was used to assess the A. baumannii biofilm formation based on the sensibility of tigecycline. The area under the ROC curve (AUC) for the biofilm formation was 0.92 (95% CI 0.864–0.981), optimal cut-off level was 1.13, sensitivity was 90.2%, and specificity was 82.1% (Figure 5).
ERIC-PCR HOMOLOGY ANALYSIS:
Cluster analysis was performed using Bionumics software, as shown in Figure 6. Correlation coefficient similarity of 0.5–0.8 is moderately correlated, and similarity of 0.80–1 is highly correlated. A total of 23 A. baumannii isolates, composed of the TS-AN group (G1, n=12) and TR-AN group (G2, n=11), could be clustered into 5 classes (ET1–ET5). It was suggested that some of the collected specimens may have come from the same clone strain and there was clonal transmission in the hospital. The drug resistance of the same clone strain to antibiotics was different, which may have been related to the different antibiotic selection pressure of the clone strain in different patients.
Discussion
Acinetobacter ssp. are oxidase-negative non-fermenting sugar gram-negative pathogenic bacteria [27,28]. Currently, the genus can be divided into 31 species according to genome, and 17 species have been named, of which
A total of 38 strains (38/72, 52.8%) isolated in this study were derived from respiratory tract specimens, indicating that
Through this study, we revealed the resistance of
In recent years, researchers have found that biofilm, which can enhance adherence to equipment and cause disease, is an important virulence factor of
Some limitations should be considered in this study. First, the association mechanism between biofilm-associated gene bfs and extracellular pump gene AdeB has yet to be further explored. Some studies have shown that inhibiting carbapenem-resistant
Conclusions
By detecting the efflux pump gene of multidrug-resistant
Figures
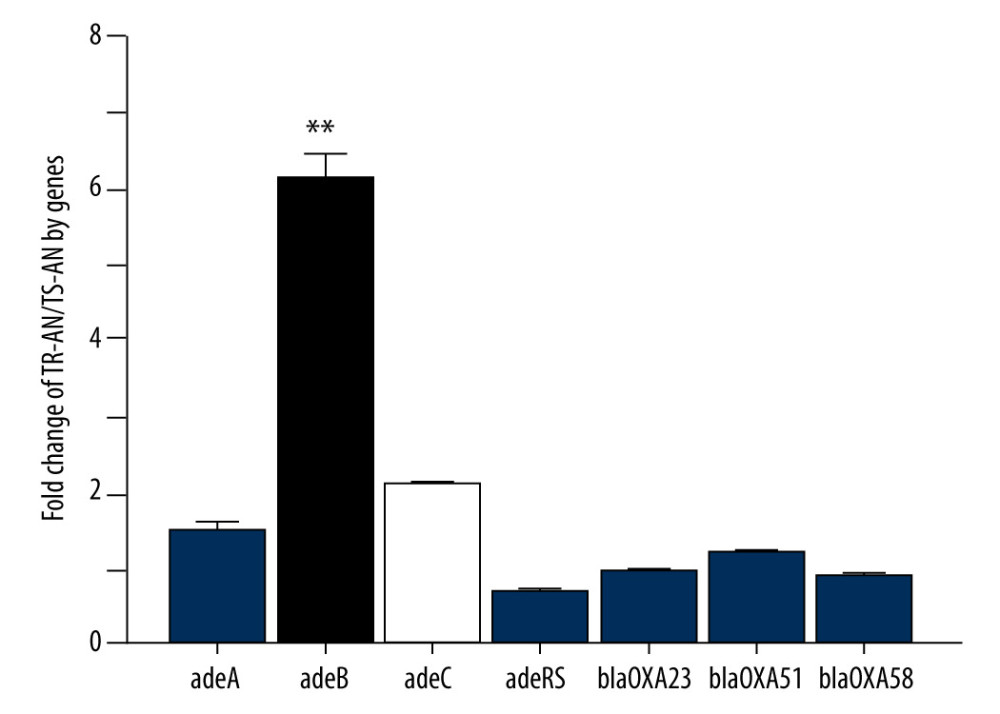 Figure 1. Relative expression of efflux pump AdeB gene in clinically isolated Acinetobacter baumannii strainsGene expression relative to that of rpoB was determined by RT-PCR. Each isolate was tested in triplicate in 2 independent experiments – white bars, TS-AN (sensitive) isolates; black bars, TR-AN (resistant) isolates. The bars represent the average, and the error bars represent the standard deviations. Data were analyzed using an independent t test (* P<0.05, ** P<0.01).
Figure 1. Relative expression of efflux pump AdeB gene in clinically isolated Acinetobacter baumannii strainsGene expression relative to that of rpoB was determined by RT-PCR. Each isolate was tested in triplicate in 2 independent experiments – white bars, TS-AN (sensitive) isolates; black bars, TR-AN (resistant) isolates. The bars represent the average, and the error bars represent the standard deviations. Data were analyzed using an independent t test (* P<0.05, ** P<0.01).  Figure 2. PCR electrophoretogram of drug resistance gene detection.
Figure 2. PCR electrophoretogram of drug resistance gene detection. 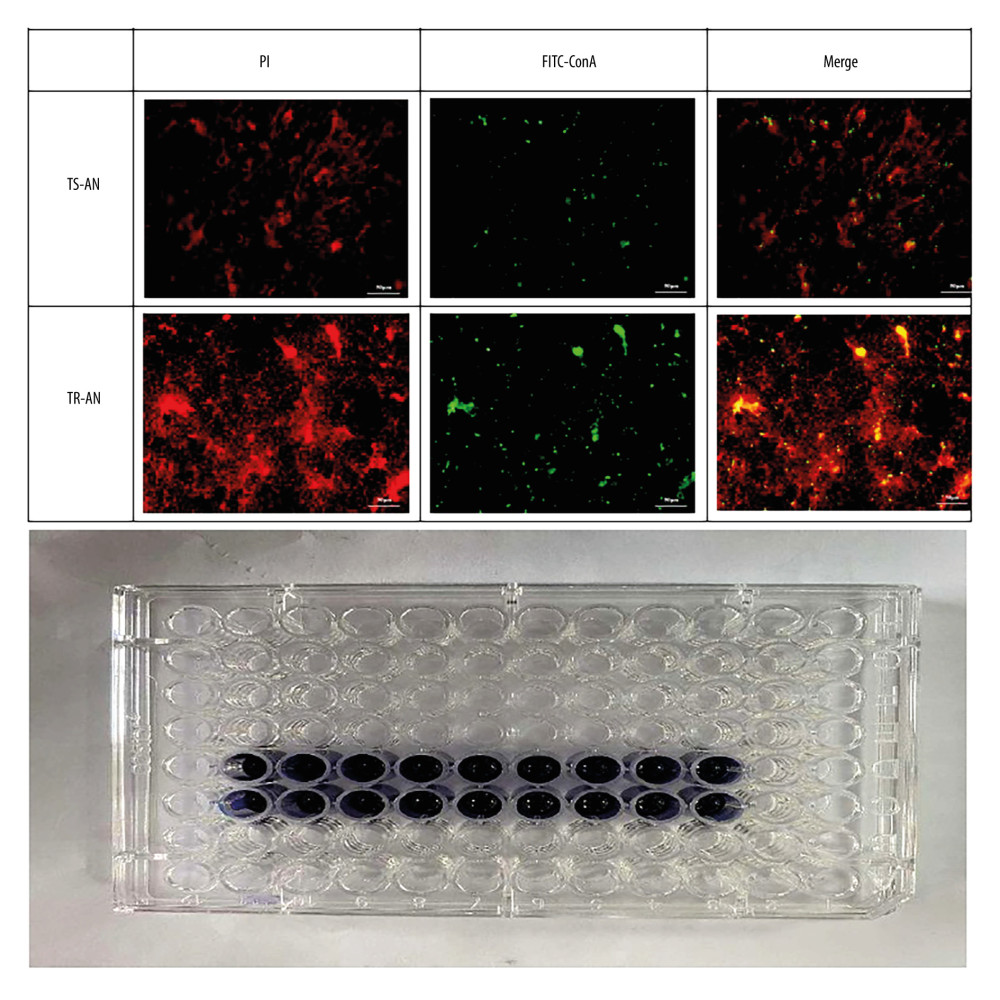 Figure 3. Observation of biofilm growth by laser confocal microscope staining diagram with crystal violet.
Figure 3. Observation of biofilm growth by laser confocal microscope staining diagram with crystal violet. 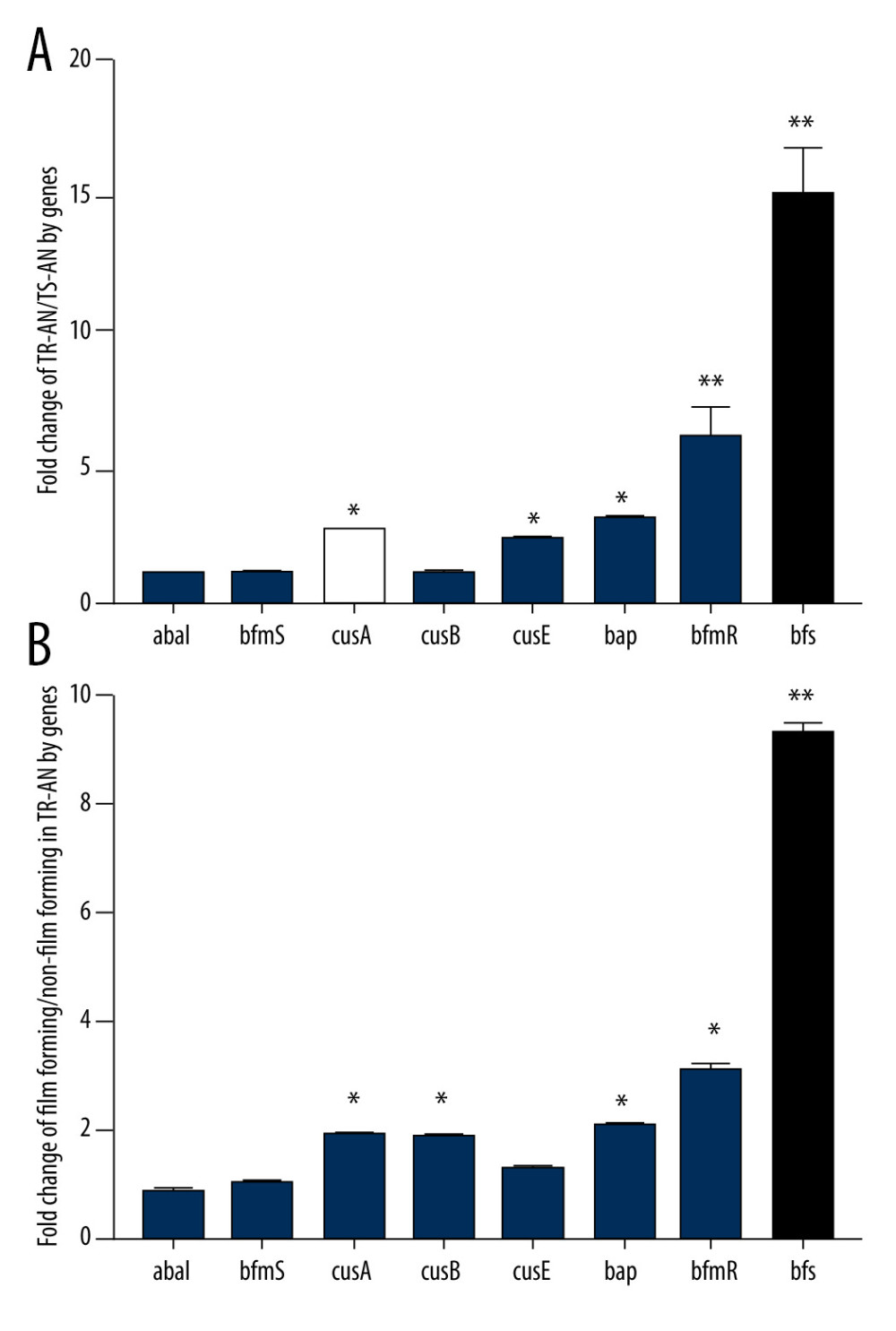 Figure 4. (A, B) Relative expression of biofilm synthesis gene bfs in clinically isolated Acinetobacter baumannii strainsGene expression relative to that of rpoB was determined by RT-PCR. Each isolate was tested in triplicate in 2 independent experiments – white bars, TS-AN (sensitive) isolates; black bars, TR-AN (resistant) isolates. The bars represent the average, and the error bars represent the standard deviations. Data were analyzed using an independent t test (* P<0.05, ** P<0.01).
Figure 4. (A, B) Relative expression of biofilm synthesis gene bfs in clinically isolated Acinetobacter baumannii strainsGene expression relative to that of rpoB was determined by RT-PCR. Each isolate was tested in triplicate in 2 independent experiments – white bars, TS-AN (sensitive) isolates; black bars, TR-AN (resistant) isolates. The bars represent the average, and the error bars represent the standard deviations. Data were analyzed using an independent t test (* P<0.05, ** P<0.01). 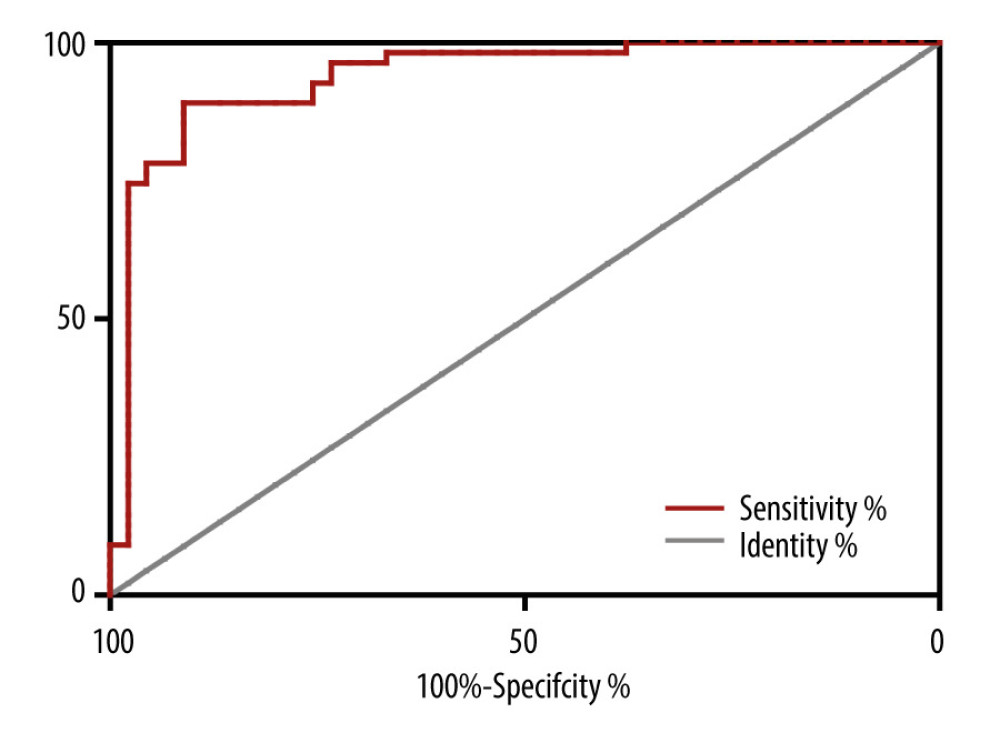 Figure 5. Receiver operating characteristic (ROC) curve for prediction of tigecycline resistance based on biofilm formation abilityOptimal cut-off levels for biofilm-forming rate were applied with ROC curves for tigecycline susceptibility. The area under the ROC curve (AUC) for the biofilm formation was 0.92 (95% CI 0.864–0.981), optimal cut-off level was 1.13, sensitivity was 90.2%, and specificity was 82.1%.
Figure 5. Receiver operating characteristic (ROC) curve for prediction of tigecycline resistance based on biofilm formation abilityOptimal cut-off levels for biofilm-forming rate were applied with ROC curves for tigecycline susceptibility. The area under the ROC curve (AUC) for the biofilm formation was 0.92 (95% CI 0.864–0.981), optimal cut-off level was 1.13, sensitivity was 90.2%, and specificity was 82.1%. 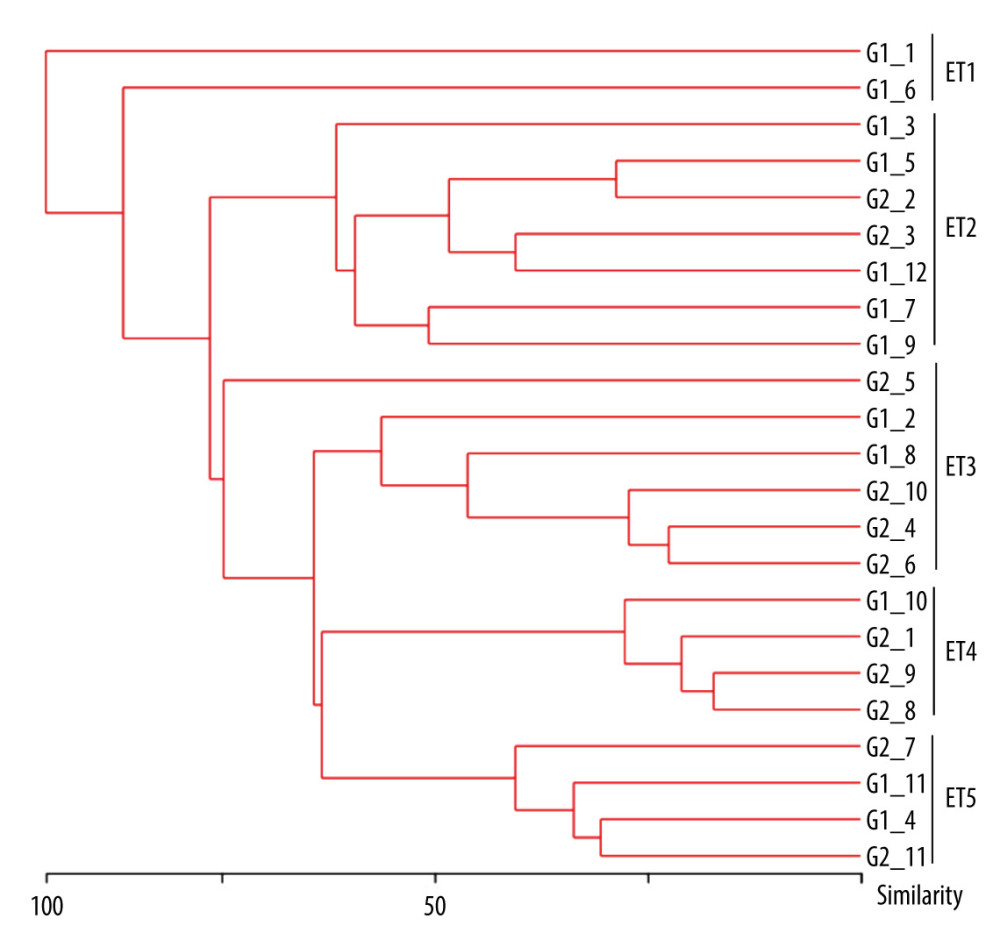 Figure 6. Dendrogram generated from ERIC-PCR banding pattern of 23 Acinetobacter baumannii isolates. The similarity analysis was performed with bionumerics software.
Figure 6. Dendrogram generated from ERIC-PCR banding pattern of 23 Acinetobacter baumannii isolates. The similarity analysis was performed with bionumerics software. Tables
Table 1. Target gene PCR primer sequences and product lengths.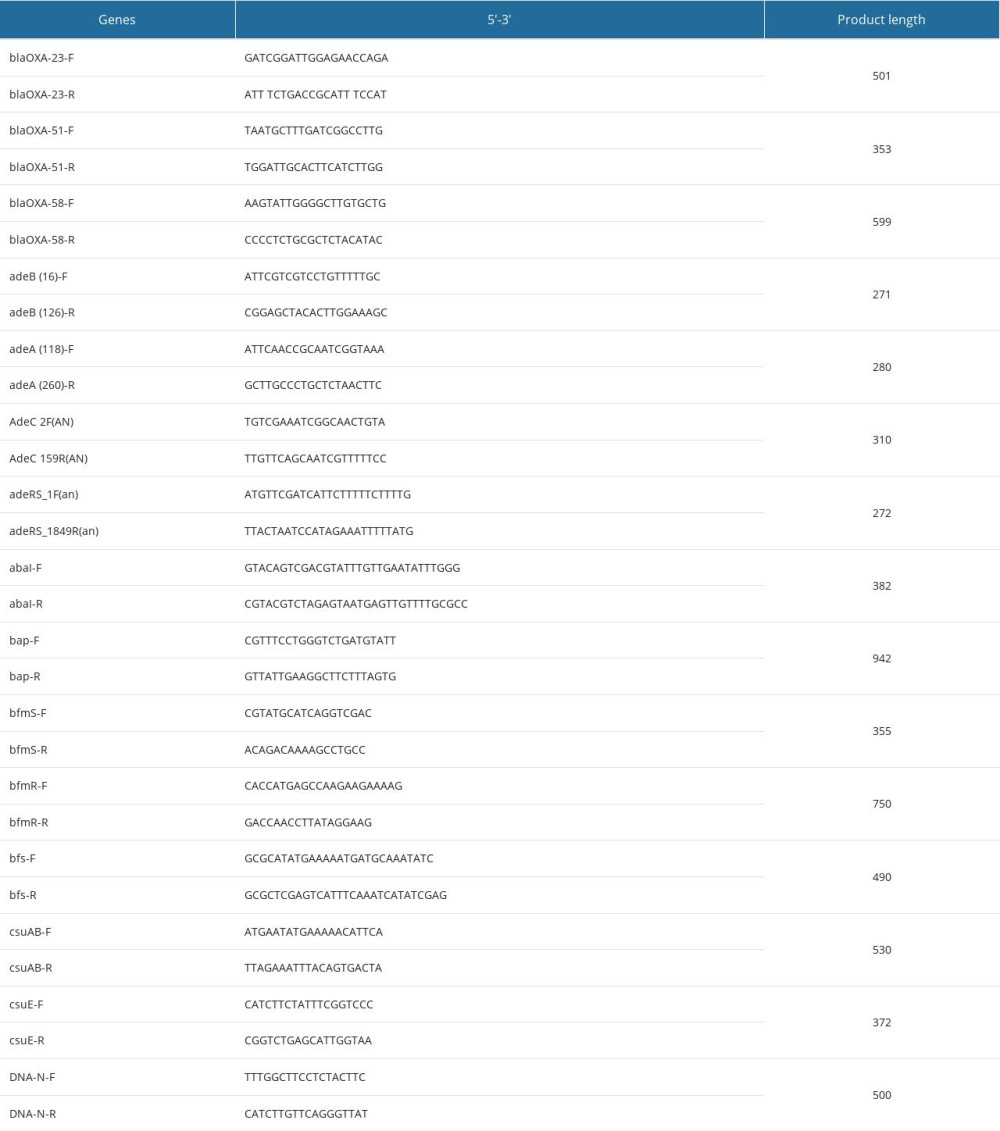 Table 2. Baseline characteristics of patients with Acinetobacter baumannii infections.
Table 2. Baseline characteristics of patients with Acinetobacter baumannii infections.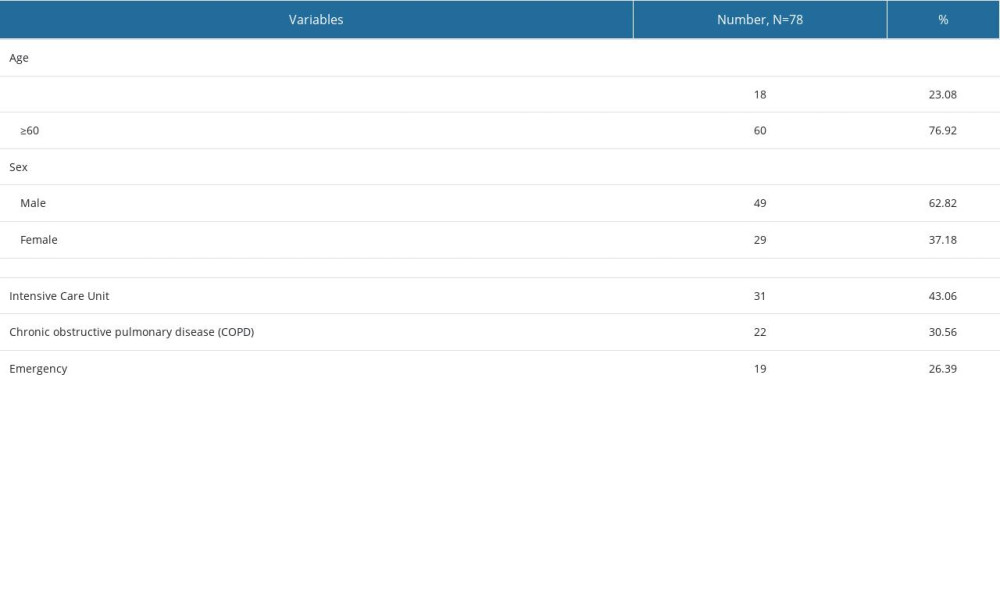 Table 3. Tigecycline minimum inhibitory concentration and expression of efflux pump genes in 72 Acinetobacter baumannii isolates.
Table 3. Tigecycline minimum inhibitory concentration and expression of efflux pump genes in 72 Acinetobacter baumannii isolates.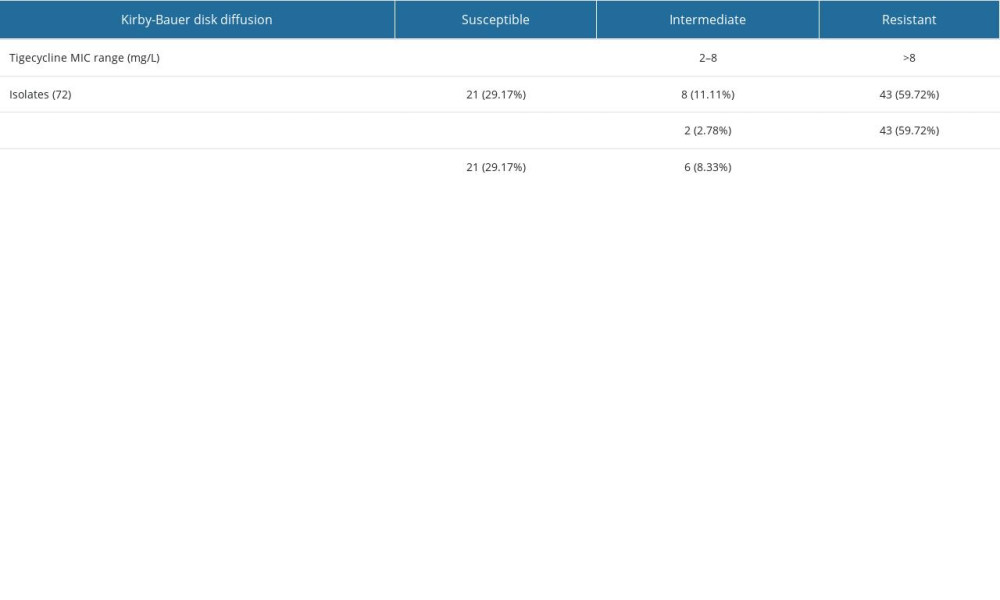 Table 4. Biofilm-related genes transcription levels in 72 Acinetobacter baumannii isolates.
Table 4. Biofilm-related genes transcription levels in 72 Acinetobacter baumannii isolates.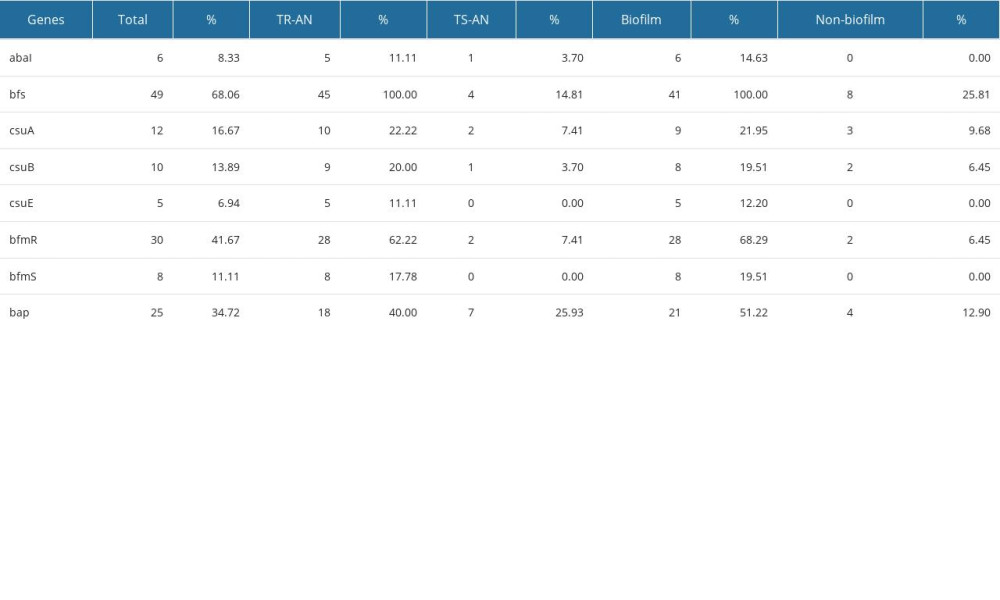 Table 5. Comparison of the ability of biofilm formation between the TR-AN (resistant) group and TS-AN (sensitive) group.
Table 5. Comparison of the ability of biofilm formation between the TR-AN (resistant) group and TS-AN (sensitive) group.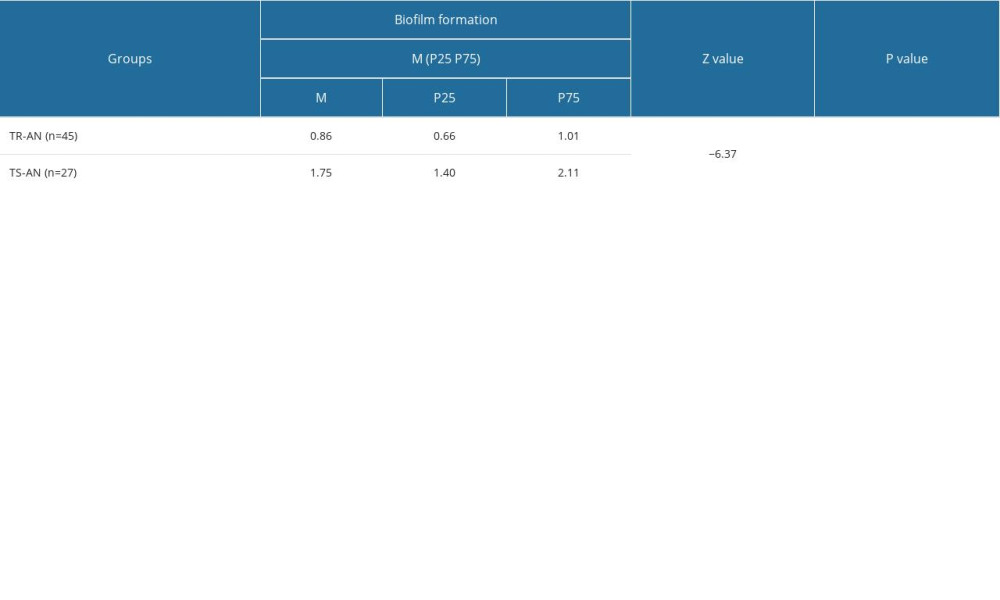
References
1. Katip W, Uitrakul S, Oberdorfer P: Pharmaceutics, 2021; 14(1); 31
2. Wienhold SM, Brack MC, Nouailles G: Viruses, 2021; 14(1); 33
3. Gedefie A, Demsis W, Ashagrie M: Infect Drug Resist, 2021; 14; 3711-19
4. Sun X, Xiang J: Curr Microbiol, 2021; 78(11); 3936-44
5. Zhang Y, Fan B, Luo Y: BMC Infect Dis, 2021; 21(1); 841, doi: 10.1186/s12.879-021-06529-2 published Online First: 20210820
6. Eze EC, Chenia HY, El Zowalaty ME: Infect Drug Resist, 2018; 11; 2277-99
7. Greene C, Wu J, Rickard AH, Xi C: Lett Appl Microbiol, 2016; 63(4); 233-39
8. Cheng Q, Cheung Y, Liu C, Structural and mechanistic basis of the high catalytic activity of monooxygenase Tet(X4) on tigecycline: BMC Biol, 2021; 19(1); 262
9. Yin T, Lai JJ, Huang WC: J Chemother, 2022; 34(3); 166-72
10. Cai L, Kong L, Wu C, Pharmacokinetics of tigecycline in both plasma and sputum in patients with severe pneumonia: J Glob Antimicrob Resist, 2021; 26; 1-3
11. Chi W, Lee HJ, Chong YP, Comparison of prospective and retrospective methods of a tigecycline post-marketing surveillance study in the safety outcomes of patients with complicated skin structure infection, complicated intraabdominal infection and community-acquired pneumonia: Infect Dis Ther, 2021; 10(1); 411-20
12. Jo J, Ko KS: Microbiol Spectr, 2021; 9(2); e0101021
13. Navidifar T, Amin M, Rashno M: Infect Drug Resist, 2019; 12; 1099-111
14. Wang YC, Kuo SC, Yang YS: Antimicrob Agents Chemother, 2016; 60(8); 4670-76
15. Bankan N, Koka F, Vijayaraghavan R: Antibiotics (Basel), 2021; 10(9); 1037
16. Li H, Wang X, Zhang Y: Future Microbiol, 2015; 10(3); 337-46
17. Ruzin A, Keeney D, Bradford PA: J Antimicrob Chemother, 2007; 59(5); 1001-4
18. Sun JR, Perng CL, Lin JC: Eur J Clin Microbiol Infect Dis, 2014; 33(12); 2141-47
19. Wang C, Huang Z, Li W, Can metagenomic next-generation sequencing identify the pathogens responsible for culture-negative prosthetic joint infection?: BMC Infect Dis, 2020; 20(1); 253
20. Yin Y, Yao H, Doijad S: Nat Commun, 2019; 10(1); 4283
21. Dalyan Cilo B, Ener B, Comparison of Clinical Laboratory Standards Institute (CLSI) microdilution method and VITEK 2 automated antifungal susceptibility system for the determination of antifungal susceptibility of Candida species: Cureus, 2021; 13(12); e20220
22. Humphries R, Bobenchik AM, Hindler JA, Schuetz AN: J Clin Microbiol, 2021; 59(12); e0021321
23. Tsala M, Vourli S, Daikos GL: J Antimicrob Chemother, 2017; 72(1); 172-80
24. Lin JN, Lai CH, Yang CH: Sci Rep, 2019; 9(1); 2267
25. Wakasugi N, Tomita T, Kondo K, Differences of fertility in reciprocal crosses between inbred strains of mice. DDK, KK and NC: J Reprod Fertil, 1967; 13(1); 41-50
26. Bayram Y, Guducuoglu H, Otlu B, Epidemiological characteristics and molecular typing of Salmonella enterica serovar Typhi during a waterborne outbreak in Eastern Anatolia: Ann Trop Med Parasitol, 2011; 105(5); 359-65
27. May HC, Yu JJ, Shrihari S: Front Microbiol, 2019; 10; 2849
28. Sykes EME, Deo S, Kumar A: Front Genet, 2020; 11; 601380
29. Ifa IA, Paul SK, Hossain MA, Isolation of Acinetobacter species from clinical specimens with detection of their antimicrobial susceptibility pattern from a tertiary care hospital, Bangladesh: Mymensingh Med J, 2020; 29(3); 622-27
30. Ioannou P, Mavrikaki V, Kofteridis DP, Infective endocarditis by Acinetobacter species: A systematic review: J Chemother, 2021; 33(4); 203-15
31. Beheshti M, Ardebili A, Beheshti F: Rev Inst Med Trop Sao Paulo, 2020; 62; e88
32. Ribeiro EA, Gales AC, Oliveira APS: Rev Soc Bras Med Trop, 2020; 54; e20200087
33. Elbehiry A, Marzouk E, Moussa IM: Saudi J Biol Sci, 2021; 28(1); 1158-66
34. Moehario LH, Esterita T, Shirleen V: J Infect Dev Ctries, 2020; 14(12); 1455-60
35. Baraka A, Traglia GM, Montana S: Antibiotics (Basel), 2020; 10(1); 16
36. Govender R, Amoah ID, Kumari S, Detection of multidrug resistant environmental isolates of acinetobacter and Stenotrophomonas maltophilia: A possible threat for community acquired infections?: J Environ Sci Health A Tox Hazard Subst Environ Eng, 2021; 56(2); 213-25
37. Weinberg SE, Villedieu A, Bagdasarian N: Infect Prev Pract, 2020; 2(3); 100077
38. Ben-Chetrit E, Wiener-Well Y, Lesho E: Crit Care, 2018; 22(1); 319
39. Jain M, Sharma A, Sen MK, Rani V: Microb Pathog, 2019; 128; 75-81
40. Ren J, Li X, Wang L: Open Med (Wars), 2019; 14; 772-77
41. Lynch JP, Zhanel GG, Clark NM: Semin Respir Crit Care Med, 2017; 38(3); 311-25
42. Khalil MAF, Ahmed FA, Elkhateeb AF: Microorganisms, 2021; 9(11); 2365
43. Amin M, Navidifar T, Shooshtari FS: Infect Drug Resist, 2019; 12; 3867-81
44. Yang CH, Su PW, Moi SH, Chuang LY: Molecules, 2019; 24(10); 1849
45. Bahador A, Farshadzadeh Z, Raoofian R: Ann Clin Microbiol Antimicrob, 2018; 17(1); 24
46. Alamri AM, Alsultan AA, Ansari MA, Alnimr AM: Pathogens, 2020; 9(8); 630
47. Chen L, Li H, Wen H: Microbiologyopen, 2020; 9(9); e1063
48. Lin MF, Lin YY, Lan CY: Peer J, 2020; 8; e9020
49. Espinal P, Marti S, Vila J: J Hosp Infect, 2012; 80(1); 56-60
50. Peng LH, Liang X, Chang RH: Biofouling, 2020; 36(7); 753-65
51. Zhang L, Ding G, Wei L: Diagn Mol Pathol, 2011; 20(4); 242-48
52. Devanga Ragupathi NK, Muthuirulandi Sethuvel DP, Triplicane Dwarakanathan H: Front Microbiol, 2020; 11; 591679
53. Rajivgandhi GN, Alharbi NS, Kadaikunnan S: Saudi J Biol Sci, 2021; 28(3); 1750-56
54. Chiu SK, Chan MC, Huang LY: PLoS One, 2017; 12(4); e0175140
Figures
 Figure 1. Relative expression of efflux pump AdeB gene in clinically isolated Acinetobacter baumannii strainsGene expression relative to that of rpoB was determined by RT-PCR. Each isolate was tested in triplicate in 2 independent experiments – white bars, TS-AN (sensitive) isolates; black bars, TR-AN (resistant) isolates. The bars represent the average, and the error bars represent the standard deviations. Data were analyzed using an independent t test (* P<0.05, ** P<0.01).
Figure 1. Relative expression of efflux pump AdeB gene in clinically isolated Acinetobacter baumannii strainsGene expression relative to that of rpoB was determined by RT-PCR. Each isolate was tested in triplicate in 2 independent experiments – white bars, TS-AN (sensitive) isolates; black bars, TR-AN (resistant) isolates. The bars represent the average, and the error bars represent the standard deviations. Data were analyzed using an independent t test (* P<0.05, ** P<0.01). Figure 2. PCR electrophoretogram of drug resistance gene detection.
Figure 2. PCR electrophoretogram of drug resistance gene detection. Figure 3. Observation of biofilm growth by laser confocal microscope staining diagram with crystal violet.
Figure 3. Observation of biofilm growth by laser confocal microscope staining diagram with crystal violet. Figure 4. (A, B) Relative expression of biofilm synthesis gene bfs in clinically isolated Acinetobacter baumannii strainsGene expression relative to that of rpoB was determined by RT-PCR. Each isolate was tested in triplicate in 2 independent experiments – white bars, TS-AN (sensitive) isolates; black bars, TR-AN (resistant) isolates. The bars represent the average, and the error bars represent the standard deviations. Data were analyzed using an independent t test (* P<0.05, ** P<0.01).
Figure 4. (A, B) Relative expression of biofilm synthesis gene bfs in clinically isolated Acinetobacter baumannii strainsGene expression relative to that of rpoB was determined by RT-PCR. Each isolate was tested in triplicate in 2 independent experiments – white bars, TS-AN (sensitive) isolates; black bars, TR-AN (resistant) isolates. The bars represent the average, and the error bars represent the standard deviations. Data were analyzed using an independent t test (* P<0.05, ** P<0.01). Figure 5. Receiver operating characteristic (ROC) curve for prediction of tigecycline resistance based on biofilm formation abilityOptimal cut-off levels for biofilm-forming rate were applied with ROC curves for tigecycline susceptibility. The area under the ROC curve (AUC) for the biofilm formation was 0.92 (95% CI 0.864–0.981), optimal cut-off level was 1.13, sensitivity was 90.2%, and specificity was 82.1%.
Figure 5. Receiver operating characteristic (ROC) curve for prediction of tigecycline resistance based on biofilm formation abilityOptimal cut-off levels for biofilm-forming rate were applied with ROC curves for tigecycline susceptibility. The area under the ROC curve (AUC) for the biofilm formation was 0.92 (95% CI 0.864–0.981), optimal cut-off level was 1.13, sensitivity was 90.2%, and specificity was 82.1%. Figure 6. Dendrogram generated from ERIC-PCR banding pattern of 23 Acinetobacter baumannii isolates. The similarity analysis was performed with bionumerics software.
Figure 6. Dendrogram generated from ERIC-PCR banding pattern of 23 Acinetobacter baumannii isolates. The similarity analysis was performed with bionumerics software. Tables
 Table 1. Target gene PCR primer sequences and product lengths.
Table 1. Target gene PCR primer sequences and product lengths. Table 2. Baseline characteristics of patients with Acinetobacter baumannii infections.
Table 2. Baseline characteristics of patients with Acinetobacter baumannii infections. Table 3. Tigecycline minimum inhibitory concentration and expression of efflux pump genes in 72 Acinetobacter baumannii isolates.
Table 3. Tigecycline minimum inhibitory concentration and expression of efflux pump genes in 72 Acinetobacter baumannii isolates. Table 4. Biofilm-related genes transcription levels in 72 Acinetobacter baumannii isolates.
Table 4. Biofilm-related genes transcription levels in 72 Acinetobacter baumannii isolates. Table 5. Comparison of the ability of biofilm formation between the TR-AN (resistant) group and TS-AN (sensitive) group.
Table 5. Comparison of the ability of biofilm formation between the TR-AN (resistant) group and TS-AN (sensitive) group. Table 1. Target gene PCR primer sequences and product lengths.
Table 1. Target gene PCR primer sequences and product lengths. Table 2. Baseline characteristics of patients with Acinetobacter baumannii infections.
Table 2. Baseline characteristics of patients with Acinetobacter baumannii infections. Table 3. Tigecycline minimum inhibitory concentration and expression of efflux pump genes in 72 Acinetobacter baumannii isolates.
Table 3. Tigecycline minimum inhibitory concentration and expression of efflux pump genes in 72 Acinetobacter baumannii isolates. Table 4. Biofilm-related genes transcription levels in 72 Acinetobacter baumannii isolates.
Table 4. Biofilm-related genes transcription levels in 72 Acinetobacter baumannii isolates. Table 5. Comparison of the ability of biofilm formation between the TR-AN (resistant) group and TS-AN (sensitive) group.
Table 5. Comparison of the ability of biofilm formation between the TR-AN (resistant) group and TS-AN (sensitive) group. In Press
12 Mar 2024 : Clinical Research
Metabolomic Alterations in Methotrexate Treatment of Moderate-to-Severe PsoriasisMed Sci Monit In Press; DOI: 10.12659/MSM.943360
14 Mar 2024 : Clinical Research
Renal Dysfunction Increases Risk of Adverse Cardiovascular Events in 5-Year Follow-Up Study of Intermediate...Med Sci Monit In Press; DOI: 10.12659/MSM.943956
15 Mar 2024 : Clinical Research
Impact of One-Lung Ventilation on Oxygenation and Ventilation Time in Thoracoscopic Heart Surgery: A Compar...Med Sci Monit In Press; DOI: 10.12659/MSM.943089
14 Mar 2024 : Clinical Research
Differential DHA and EPA Levels in Women with Preterm and Term Births: A Tertiary Hospital Study in IndonesiaMed Sci Monit In Press; DOI: 10.12659/MSM.943895
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952








