20 June 2020: Database Analysis
Special AT-rich Sequence Binding-Protein 1 (SATB1) Correlates with Immune Infiltration in Breast, Head and Neck, and Prostate Cancer
Hua Ge1ABCDEF*, Yan Yan2BCDE, Maozhao Yan1CEF, Lingfei Guo1EF, Kun Mao1CFDOI: 10.12659/MSM.923208
Med Sci Monit 2020; 26:e923208
Abstract
BACKGROUND: SATB1 is essential in gene regulation and associates with T cell development. Aberrant SATB1 expression has been reported in various neoplasms. However, correlations between SATB1 and tumor immune infiltration and prognosis in malignancies still remains unclear.
MATERIAL AND METHODS: We used Oncomine and the Tumor Immune Estimation Resource database to explore the expression of SATB1 in cancers. In addition, Kaplan-Meier plotter, PrognoScan, and Gene Expression Profiling Interactive Analysis were also used to assess the effects of SATB1 on clinical prognosis. Furthermore, correlations between cancer immune infiltration and SATB1 were analyzed via Tumor Immune Estimation Resource.
RESULTS: The results demonstrated that SATB1 correlates with prognosis in different types of cancers, such as breast invasive carcinoma (BRAC), head and neck cancer (HNSC), and prostate adenocarcinoma (PRAD). Decreased expression of SATB1 was associated with poor overall and progression-free survival of BRAC patients with positive estrogen receptor (ER) as well as mutated TP53. In addition, B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells infiltration in BRAC, HNSC, and PRAD were also correlated with SATB1 expression level. Moreover, we found strong correlations between SATB1 and various immune markers for BRAC, HNSC, and PRAD.
CONCLUSIONS: In BRAC, HNSC, and PRAD patients, SATB1 has potential to serve as a prognostic indicator for predicting tumor immune infiltration and prognosis.
Keywords: Breast Neoplasms, Head and Neck Neoplasms, Prostatic Neoplasms, Tumor Escape, B-Lymphocytes, CD4-Positive T-Lymphocytes, CD8-Positive T-Lymphocytes, Databases, Factual, Databases, Genetic, Lymphocytes, Tumor-Infiltrating, Macrophages, Matrix Attachment Region Binding Proteins, Neutrophils, RNA, Messenger, Squamous Cell Carcinoma of Head and Neck
Background
Breast invasive carcinoma (BRCA), head and neck cancer (HNSC), and prostate adenocarcinoma (PRAD) are common malignancies in the world, and they remain a major public health problem [1]. The immune infiltrate mechanism is involved in the development of cancers, and immunotherapy has been proven to be a promising strategy in BRCA, HNSC, and PRAD [2–4]. In BRCA, human epidermal growth factor receptor 2 (HER2) is partially mediated by immune mechanisms and HER2 positive patients respond well to HER2 targeted therapy [5]. For HNSC patients with recurrence and metastases, anti-PD1 antibodies can improve the overall survival (OS) [6,7]. In addition, immuno-therapeutic medicine such as anti-PD1 and anti-CTLA4 show a partial response in PRAD patients [8]. Studies also have indicated that tumor infiltrating neutrophils (TINs) and tumor associated macrophages (TAMs) have effects on survival and therapeutic efficacy in cancer patients [9,10]. Therefore, exploring novel immune-related therapeutic targets is important for facilitating individualization and optimization of cancer patients in treatment.
The special AT-rich sequence binding-protein 1 (SATB1) is one kind of protein that binds to nuclear matrix. SATB1 participates in the mechanisms of chromatin remodeling and regulates gene expression [11]. SATB1 can activate or repress genes by interacting with the PDZ domain of chromatin modifying enzymes [12]. Abnormal expression of SATB1 has been reported in different neoplasms such as glioma, nasopharyngeal, breast, lung, pancreatic, liver, colorectal, kidney, bladder, prostate, ovarian, lymphoma, and so on [13]. SATB1 is essential in T cell maturation and is correlated with thymocyte development and T-helper 2 (Th2) cell activation [14]. A recent study suggested that SATB1 regulates PDCD1 expression during T cell activation and prevents T cell exhaustion, and that dysregulation of this pathway results in anti-tumor immune dysfunction [15]. These findings reveal that SATB1 plays vital roles in tumor infiltrating lymphocytes. However, the specific mechanism of SATB1 in the regulation of tumor immunity remains unclear.
In this study, Oncomine, PrognoScan, Kaplan-Meier plotter (K-M plot), and Gene Expression Profiling Interactive Analysis (GEPIA) were employed to estimate the correlation between the prognosis of cancer patients and SATB1. Additionally, we explored SATB1 expression in immune infiltrating cells in multiple tumors through the Tumor Immune Estimation Resource (TIMER) database. Our results provided insight on SATB1 in tumor-immune interactions in BRCA, HNSC, and PRAD.
Material and Methods
ONCOMINE ANALYSIS:
We used Oncomine (https://www.oncomine.org/) and the TIMER site (https://cistrome.shinyapps.io/timer/) to assess the expression of SATB1 in different cancers [16,17]. The threshold was as follows: fold change=1.5, P-value=0.001.
PROGNOSCAN ANALYSIS:
PrognoScan (http://www.abren.net/PrognoScan/) was employed to assess the correlation between SATB1 and survival in cancer patients [18]. Cox P-value <0.05 was considered to be statistically significant.
K-M PLOT ANALYSIS:
The K-M plot (http://kmplot.com/) is a website which can evaluate the survival of breast, ovarian, lung, and gastric cancer patient samples [19]. Thus, we explored the effect of SATB1 on prognosis in breast, ovarian, lung, and gastric cancer patients via K-M plot.
GEPIA ANALYSIS:
Among 33 different types of cancer, GEPIA (
TIMER ANALYSIS:
TIMER is a website tool based on TCGA, which includes 10 897 samples across 32 cancers to evaluate tumor immune infiltration [17]. We analyzed the association between SATB1 and immune infiltration cells, including B cells, CD4+ T cells, CD8+ T cells, macrophages, neutrophils, and dendritic cells (DCs). We looked at the expression level of SATB1 versus tumor purity [20]. Additionally, we also estimated the correlation between SATB1 and biomarkers of tumor infiltrating cells, including markers of CD8+ T cells, T cells (general), B cells, monocytes, TAMs, M1 and M2 macrophages, neutrophils, natural killer (NK) cells, DCs, T-helper 1 (Th1) and Th2 cells, follicular helper T (Tfh) cells, T-helper 17 (Th17) cells, regulatory T cells (Tregs), and exhausted T cells [21]. TIMER can generate scatter plots as well as Spearman’s correlation and P-value. The SATB1 gene symbol is displayed on the x-axis, and correlated gene symbols are shown on the y-axis. The expression level of genes was adjusted to log2 RSEM. A flow chart of the study design is shown in Figure 1.
STATISTICAL ANALYSIS:
Survival curves were produced via PrognoScan and K-M plot. The results of Oncomine are shown as
Results
SATB1 EXPRESSION IN VARIOUS CANCERS:
As revealed in Figure 2A, the results from the Oncomine database indicated that the expression of SATB1 was lower in brain, breast, colorectal, head and neck, lung, leukemia, lymphoma, liver, ovarian, melanoma, and sarcoma carcinomas, while SATB1 expression was increased in leukemia and myeloma tissues in some data sets.
To investigate the expression of SATB1 in malignancy, we used RNA-seq data from TCGA. As presented in Figure 2B, SATB1 expression was decreased in bladder urothelial carcinoma (BLCA), BRCA, cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), HNSC, kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), PRAD, rectum adenocarcinoma (READ), skin cutaneous melanoma (SKCM), stomach adenocarcinoma (STAD), and uterine corpus endometrial carcinoma (UCEC) tissues. However, SATB1 expression was increased in kidney chromophobe (KICH) tissues.
PROGNOSTIC VALUE OF SATB1 IN CANCERS:
The effect of SATB1 expression on survival was assessed through PrognoScan; the results are presented in Table 1. In particularly, the expression of SATB1 significantly affected prognosis in 7 types of cancers, including breast cancer (Figure 3A–3E), lung cancer (Figures 3F–3I), brain cancer (Figure 3J, 3K), prostate cancer (Figure 3L), skin cancer (Figure 3M), bladder cancer (Figure 3N), and eye cancer (Figure 3O). In addition, one cohort (GSE2990) [22] which included 125 samples of BRCA, demonstrated that decreased expression of SATB1 was associated with worse prognosis (relapse-free survival [RFS] HR: 0.52, 95% confidence interval [CI]: 0.33–0.81, Cox P: 0.004; distant metastasis free survival [DMFS] HR: 0.53, 95% CI: 0.30–0.96, Cox P=0.037).
We also used K-M plot to explore the prognostic effects of SATB1 based on Affymetrix microarrays. As shown in Figure 4A and 4B, a lower level of SATB1 was correlate with worse prognosis in BRCA patients (OS HR: 0.72, 95% CI: 0.58–0.89, P: 0.0027; RFS HR: 0.77, 95% CI: 0.66–0.90, P: 0.0012). In addition, a low level of SATB1 was correlated with poor OS in LUAD (HR: 0.57, 95% CI: 0.49–0.68) (Figure 4C). However, SATB1 had less influence on progression-free survival (PFS) (Figure 4D) in LUAD, and OS and PFS in gastric cancer and ovarian cancer (Figure 4E–4H).
The effect of SATB1 on survival in different cancers was assessed using the GEPIA database. Decreased expression of SATB1 was associated with worse OS and disease-free survival (DFS) in KIRC, LGG, SKCM, and uveal melanoma (UVM); and DFS in PRAD and OS in SARC (sarcoma). Additionally, a high level of SATB1 was associated with worse DFS in STAD (P<0.05) (Table 2). These results suggested that SATB1 has the potential to predict prognosis in multiple types of cancers.
LOW LEVEL OF SATB1 IMPACTS THE PROGNOSIS OF BRCA PATIENTS WITH POSITIVE ESTROGEN RECEPTOR (ER) AND MUTATED TP53:
To comprehensive understand the potential mechanisms of SATB1 expression in BRCA, we used the K-M plot to explore the association between SATB1 and clinical characteristics of BRCA patients. As shown in Table 3, a low level of SATB1 was associated with poor OS and PFS in BRCA patients with positive estrogen receptor (ER) and mutated TP53. Meanwhile, a low level of SATB1 was also associated with PFS in patients with HER2 negative and luminal B of intrinsic subtype, and OS in patients with grade 2 and basal-like 2 of Pietenpol subtype (P<0.05).
SATB1 WAS ASSOCIATED WITH TUMOR IMMUNE INFILTRATION IN BRCA, HNSC, AND PRAD:
We used the TIMER database to investigate the associations of SATB1 and tumor immune infiltration in 33 types of cancer. As shown Figures 5–8, the results indicate that SATB1 was significantly correlated with tumor purity and B cell infiltration in 16 and 14 types of cancer, respectively. Additionally, SATB1 expression was associated with infiltration of CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and DCs in 11, 19, 25, 16, and 18 types of cancer, respectively.
We further explored the specific cancer type in which SATB1 correlated with tumor immune infiltration. Accordingly, we found that SATB1 was negative related to tumor purity (r=−0.291, P=6.71e-21) and positive correlated with infiltration of B cells (r=0.116, P=2.83e-04), CD8+ T cells (r=0.251, P=1.89e-15), CD4+ T cells (r=0.218, P=7.86e-12), macrophages (r=0.151, P=1.95e-06), neutrophils (r=0.232, P=4.32e-13), and DCs (r=0.189, P=4.01e-09) in BRCA (Figure 5A). In addition, SATB1 was negative correlated with tumor purity (r=−0.103, P=2.17e-02) and positive correlated with infiltration of B cells (r=0.362, P=3.61e-16), CD8+ T cells (r=0.332, P=1.26e-13), CD4+ T cells (r=0.455, P=7.26e-27), macrophages (r=0.381, P=3.75e-18), neutrophils (r=0.216, P=1.73e-06), and DCs (r=0.430, P=3.97e-23) in HNSC (Figure 5B). Furthermore, SATB1 expression was negative correlated with tumor purity (r=−0.178, P=2.52e-04) and positive correlated with infiltration of B cells (r=0.195, P=6.81e-05), CD8+ T cells (r=0.382, P=6.47e-16), CD4+ T cells (r=0.104, P=3.47e-02), macrophages (r=0.281, P=5.30e-09), neutrophils (r=0.240, P=8.21e-07), and DCs (r=0.267, P=3.13e-08) in PRAD (Figure 5C). Our findings indicate that SATB1 is essential in immune cells infiltration in BRAC, HNSC, and PRAD.
CORRELATION BETWEEN SATB1 AND IMMUNE MARKER SETS:
We investigated the associations between SATB1 and immune markers of diverse immune cells in BRCA, HNSC, and PRAD through the TIMER database. After adjustments for tumor purity, our findings revealed that SATB1 was significantly related to most immune markers of different immune cells and diverse T cells in BRAC, HNSC, and PRAD (Figure 9A–9C, Table 4).
As shown in Table 4, we discovered that the expression levels of gene markers of CD8+ T cell, T cells (general), B cells, and neutrophils had significant correlations with SATB1 both in BRAC and HNSC. In addition, the expression levels of gene markers of M1 macrophages and Th2 cells had significant correlations with SATB1 both in HNSC and PRAD. We also confirmed that the expression of gene markers of monocytes, TAMs, M2 macrophages, DCs, Th2 cells, Tfh cells, and Tregs significantly correlated with SATB1 in HNSC. These findings indicated that SATB1 is critical to immune escape in BRCA, HNSC, and PRAD microenvironments.
Discussion
The recognition that immune cells can identify and destroy cancer cells has promoted a tremendous shift in the perception of cancers, and immunotherapies have been shown to have curative effects in tumors which were resistant to regular therapy [23]. SATB1 can reprogram gene expression profiles and cause rapid phenotype changes. Increasingly, studies have demonstrated that SATB1 is essential in deterioration of tumors [13]. In our research, we found that aberrant SATB1 expression was related to prognosis in diverse cancers. Lower SATB1 expression correlated with poor survival in BRCA patients with positive ER and mutated TP53. Additionally, our results indicated that in BRAC, HNSC, and PRAD, the infiltration levels of immune cell and different immune gene markers were related to SATB1 expression. Therefore, our study provides a theoretical basis for understanding the function of SATB1 in tumor progression and its application as a tumor biomarker.
In this study, datasets in Oncomine and TIMER were used to explore the expression of SATB1 and its prognostic value in human cancers. We found SATB1 was differentially expressed between cancer and normal tissues in various malignancies. Oncomine analysis revealed that the expression of SATB1 was reduced in brain, breast, colorectal, head and neck, leukemia, liver, lung, lymphoma, melanoma, ovarian, and sarcoma carcinomas, while SATB1 expression was increased in leukemia and myeloma tissues. However, our findings from TCGA data indicated that SATB1 expression was decreased in BLCA, BRCA, CHOL, COAD, ESCA, HNSC, KIRC, KIRP, LIHC, LUAD, LUSC, PRAD, READ, SKCM, STAD, and UCEC; while SATB1 expression was increased in KICH. The differences of SATB1 expression among different types of cancer in diverse databases might be due to the diversity of biological functions of SATB1 as well as data collection approaches. However, we found consistent correlations between SATB1 and prognosis in breast, colorectal, head and neck, liver, lung, gastric, and sarcoma carcinomas. GEPIA analysis based on TCGA data indicated that lower SATB1 expression was related to a worse prognosis for certain cancer types, such as KIRC, LGG, SKCM, PRAD, SARC, and UVM, while elevated SATB1 expression correlated with a better prognosis in STAD. In addition, K-M plot and PrognoScan analysis indicated decreased SATB1 expression was related to short survival in breast, lung, brain, gastric, ovarian, prostate, skin, bladder, and eye cancer patients. Moreover, depletion of SATB1 led to poor OS and PFS in BRAC patients with positive ER and mutated TP53. Thus, these results demonstrated that SATB1 could be used as a prognostic indicator in multiple types of neoplasms.
Another important finding in our research was that a low level of SATB1 was associated with different levels of immune infiltration in neoplasms, especially in BRAC, HNSC, and PRAD. Our results revealed that the SATB1 expression level had significant positive correlation with infiltration levels of B cells, CD4+ T cells, CD8+ T cells, macrophages, neutrophils, and DCs in BRAC, HNSC, and PRAD. M1 and M2 macrophage markers, such as NOS2, PTGS2, IRF5, CD163, VSIG4, and MS4A4A, showed weak to strong correlations with SATB1 expression, which indicated the regulating function of SATB1 in TAM polarization. Decreased SATB1 expression also was positively associated with the Treg and T cell exhaustion markers, such as FOXP3, CCR8, STAT5B, TGF β, and PD-1 in BRAC, HNSC and PRAD. In addition, significant correlations between SATB1 and the regulation markers of T helper cells, such as Th1, Th2, Tfh, and Th17, were found in BRAC, HNSC, and PRAD. These results suggest that SATB1 is correlated with tumor immune infiltration and plays a vital role in regulation and enrollment of tumor immune infiltrating cells in BRAC, HNSC, and PRAD.
Diverse mechanisms involved in the carcinogenesis, immune infiltration, and prognosis of SATB1 have been investigated. SATB1 has been shown to be aberrantly expressed in different types of cancers [24]. Generally, SATB1 expression has been positively related to tumor size, lymph node metastasis, tumor evolution, and prognosis in most cancer types [25–27]. A recent study reported that SATB1 regulates PDCD1 expression during T cell activation and prevents T cell exhaustion [15]. Tesone et al. reported that dynamic variations in SATB1 expression were required for the generation and immuno-stimulatory activity of conventional DCs; however, overexpressed SATB1 in differentiated DCs could convert them into pro-inflammatory or tolerogenic cells and prompt malignant transformation [28]. In an
In this study, there were several limitations. First, the cutoff value varied in the different online databases, which may introduce potential heterogeneity. Second, the number of samples in the different databases was still limited. Thus, in the future, more studies with a large number of samples are needed to provide more reliable evidence to validate the impact of SATB1 on tumor immune infiltration.
Conclusions
In summary, a low level of SATB1 expression was associated with poor survival rates and enhanced the immune infiltration level of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and DCs in various types of cancer, especially in BRAC, HNSC, and PRAD. Decreased SATB1 expression was also associated with the regulation of TAM, Treg, and T cell exhaustion in BRAC, HNSC, and PRAD. Thus, SATB1 possibly plays a vital role in enrollment and regulation of tumor immune infiltration in BRAC, HNSC, and PRAD.
Figures
 Figure 1. Flow chart of the study procedures.
Figure 1. Flow chart of the study procedures. 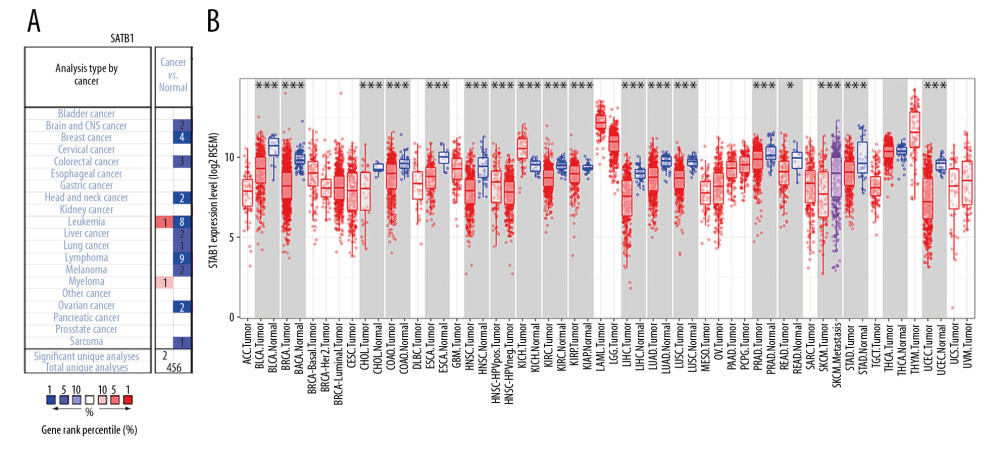 Figure 2. The expression of SATB1 in human cancers. (A) High or low SATB1 expression in data sets of different cancers in Oncomine database. (B) SATB1 expression levels in different tumor types from Tumor Immune Estimation Resource database (* P<0.05; ** P<0.01; *** P<0.001).
Figure 2. The expression of SATB1 in human cancers. (A) High or low SATB1 expression in data sets of different cancers in Oncomine database. (B) SATB1 expression levels in different tumor types from Tumor Immune Estimation Resource database (* P<0.05; ** P<0.01; *** P<0.001). ![Kaplan-Meier survival curves comparing the increased and decreased expression of SATB1 in different types of cancer in PrognoScan. (A–E) Survival curves of DSS, OS, RFS, DFS, and DMFS in breast cancer cohorts [GSE3494-GPL96 (n=236), GSE9893 (n=155), GSE2990 (n=125), and GSE4922 (n=249)]. (F–I) Survival curves of RFS, OS, and DSS in lung cancer cohorts [GSE31210 (n=204), Jacob-00182-MSK (n=104), GSE13213 (n=117), and GSE14814 (n=90)]. (J, K) Survival curves of OS in brain cancer cohorts [GSE4271-GPL96 (n=77), MGH-glioma (n=50)]. (L) Survival curves of OS in prostate cancer cohorts [GSE16560 (n=281)]. (M) Survival curves of OS in skin cancer cohorts [GSE19234 (n=38)]. (N) Survival curves of DSS in bladder cancer cohorts [GSE13507 (n=165)]. (O) Survival curves of DSS in eye cancer cohorts [GSE22138 (n=63)]. DSS – disease-specific survival; OS – overall survival; RFS – relapse-free survival; DFS – disease-free survival; DMFS – distant metastasis free survival.](https://jours.isi-science.com/imageXml.php?i=medscimonit-26-e923208-g003.jpg&idArt=923208&w=1000) Figure 3. Kaplan-Meier survival curves comparing the increased and decreased expression of SATB1 in different types of cancer in PrognoScan. (A–E) Survival curves of DSS, OS, RFS, DFS, and DMFS in breast cancer cohorts [GSE3494-GPL96 (n=236), GSE9893 (n=155), GSE2990 (n=125), and GSE4922 (n=249)]. (F–I) Survival curves of RFS, OS, and DSS in lung cancer cohorts [GSE31210 (n=204), Jacob-00182-MSK (n=104), GSE13213 (n=117), and GSE14814 (n=90)]. (J, K) Survival curves of OS in brain cancer cohorts [GSE4271-GPL96 (n=77), MGH-glioma (n=50)]. (L) Survival curves of OS in prostate cancer cohorts [GSE16560 (n=281)]. (M) Survival curves of OS in skin cancer cohorts [GSE19234 (n=38)]. (N) Survival curves of DSS in bladder cancer cohorts [GSE13507 (n=165)]. (O) Survival curves of DSS in eye cancer cohorts [GSE22138 (n=63)]. DSS – disease-specific survival; OS – overall survival; RFS – relapse-free survival; DFS – disease-free survival; DMFS – distant metastasis free survival.
Figure 3. Kaplan-Meier survival curves comparing the increased and decreased expression of SATB1 in different types of cancer in PrognoScan. (A–E) Survival curves of DSS, OS, RFS, DFS, and DMFS in breast cancer cohorts [GSE3494-GPL96 (n=236), GSE9893 (n=155), GSE2990 (n=125), and GSE4922 (n=249)]. (F–I) Survival curves of RFS, OS, and DSS in lung cancer cohorts [GSE31210 (n=204), Jacob-00182-MSK (n=104), GSE13213 (n=117), and GSE14814 (n=90)]. (J, K) Survival curves of OS in brain cancer cohorts [GSE4271-GPL96 (n=77), MGH-glioma (n=50)]. (L) Survival curves of OS in prostate cancer cohorts [GSE16560 (n=281)]. (M) Survival curves of OS in skin cancer cohorts [GSE19234 (n=38)]. (N) Survival curves of DSS in bladder cancer cohorts [GSE13507 (n=165)]. (O) Survival curves of DSS in eye cancer cohorts [GSE22138 (n=63)]. DSS – disease-specific survival; OS – overall survival; RFS – relapse-free survival; DFS – disease-free survival; DMFS – distant metastasis free survival. 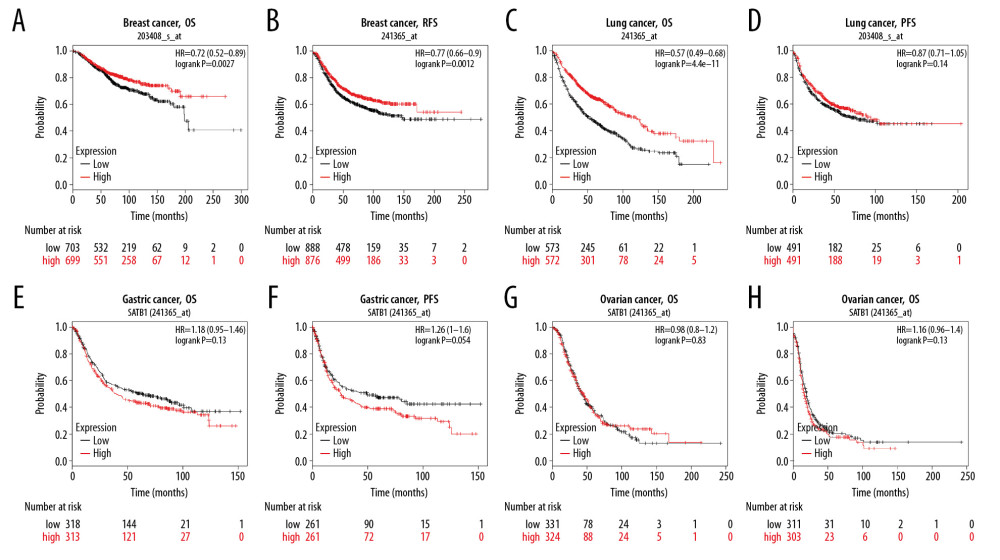 Figure 4. Kaplan-Meier survival curves comparing the expression of SATB1 in different types of cancer in Kaplan-Meier plotter database. (A, B) OS and RFS of breast cancer (n=1402, n=977). (C, D) OS and PFS of lung cancer (n=1145, n=982). (E, F) OS and PFS of gastric cancer (n 631, n=522). (G, H) OS and PFS of ovarian cancer (n=655, n=1435). OS – overall survival; RFS – relapse-free survival; PFS – progression-free survival.
Figure 4. Kaplan-Meier survival curves comparing the expression of SATB1 in different types of cancer in Kaplan-Meier plotter database. (A, B) OS and RFS of breast cancer (n=1402, n=977). (C, D) OS and PFS of lung cancer (n=1145, n=982). (E, F) OS and PFS of gastric cancer (n 631, n=522). (G, H) OS and PFS of ovarian cancer (n=655, n=1435). OS – overall survival; RFS – relapse-free survival; PFS – progression-free survival. 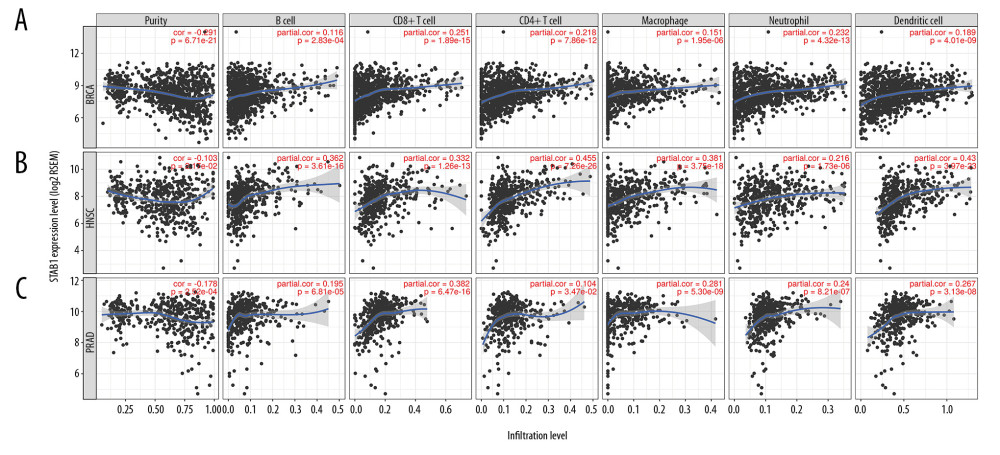 Figure 5. SATB1 expression is negatively correlated with tumor purity and positively correlated with infiltration levels of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells in BRCA, HNSC, and PRAD. (A) Correlation of SATB1 expression with immune infiltration level in BRAC (n=1093). (B) Correlation of SATB1 expression with immune infiltration level in HNSC (n=520). (C) Correlation of SATB1 expression with immune infiltration level in PRAD (n=497). BRCA – breast invasive carcinoma; HNSC – head and neck cancer; PRAD – prostate adenocarcinoma.
Figure 5. SATB1 expression is negatively correlated with tumor purity and positively correlated with infiltration levels of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells in BRCA, HNSC, and PRAD. (A) Correlation of SATB1 expression with immune infiltration level in BRAC (n=1093). (B) Correlation of SATB1 expression with immune infiltration level in HNSC (n=520). (C) Correlation of SATB1 expression with immune infiltration level in PRAD (n=497). BRCA – breast invasive carcinoma; HNSC – head and neck cancer; PRAD – prostate adenocarcinoma. 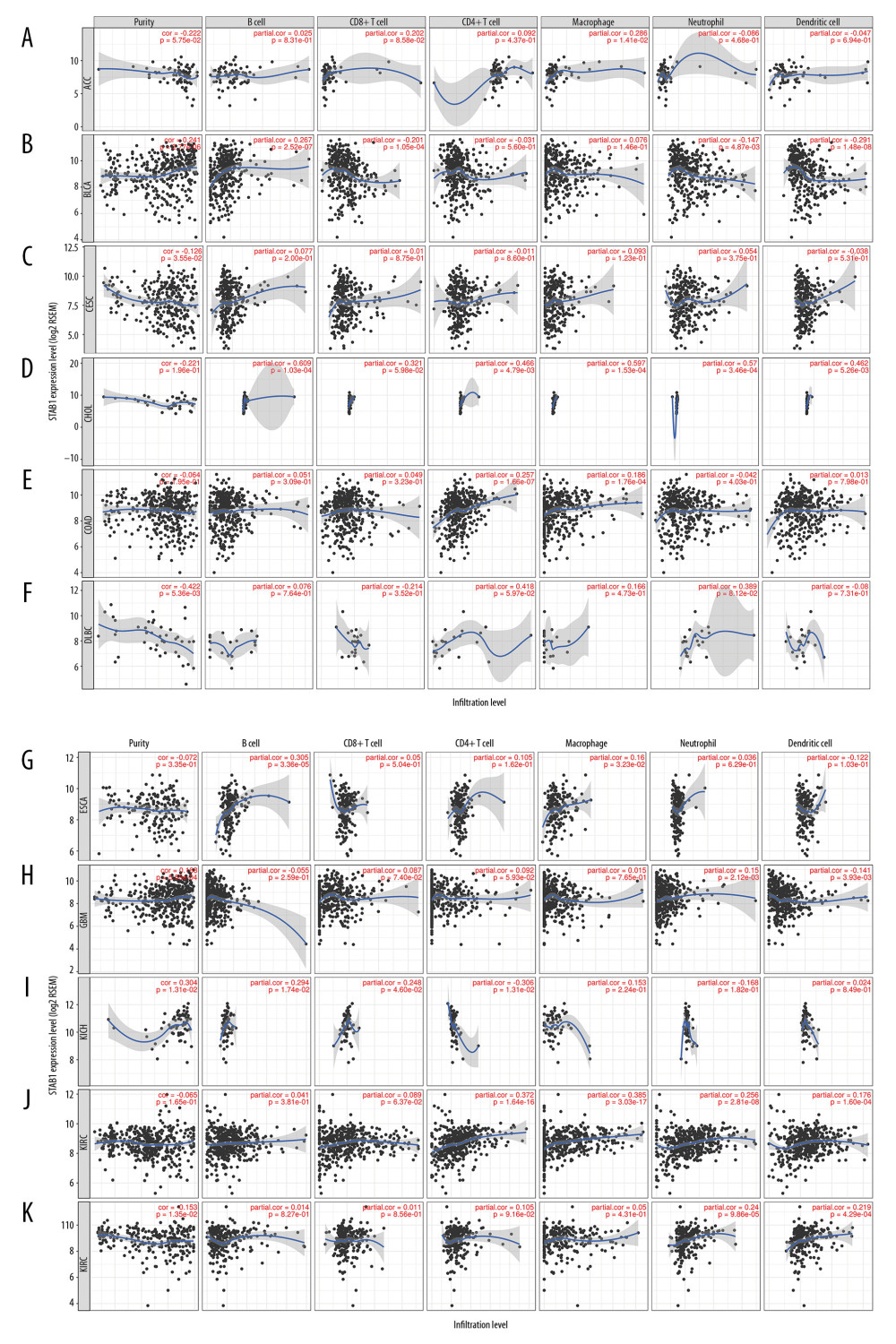 Figure 6. Association of SATB1 expression with immune infiltration levels in (A) ACC. (B) BLCA. (C) CESC. (D) CHOL. (E) COAD. (F) DLBC. (G) ESCA. (H) GBM. (I) KICH. (J) KIRC. (K) KIRP. ACC – adenoid cystic carcinoma; BLCA – bladder urothelial carcinoma; CESC – cervical squamous cell carcinoma; CHOL – cholangiocarcinoma; COAD – colon adenocarcinoma; DLBC – diffuse large B-cell lymphoma; ESCA – esophageal carcinoma; GBM – glioblastoma; KICH – kidney chromophobe; KIRC – kidney renal clear cell carcinoma; KIRP – kidney renal papillary cell carcinoma.
Figure 6. Association of SATB1 expression with immune infiltration levels in (A) ACC. (B) BLCA. (C) CESC. (D) CHOL. (E) COAD. (F) DLBC. (G) ESCA. (H) GBM. (I) KICH. (J) KIRC. (K) KIRP. ACC – adenoid cystic carcinoma; BLCA – bladder urothelial carcinoma; CESC – cervical squamous cell carcinoma; CHOL – cholangiocarcinoma; COAD – colon adenocarcinoma; DLBC – diffuse large B-cell lymphoma; ESCA – esophageal carcinoma; GBM – glioblastoma; KICH – kidney chromophobe; KIRC – kidney renal clear cell carcinoma; KIRP – kidney renal papillary cell carcinoma. 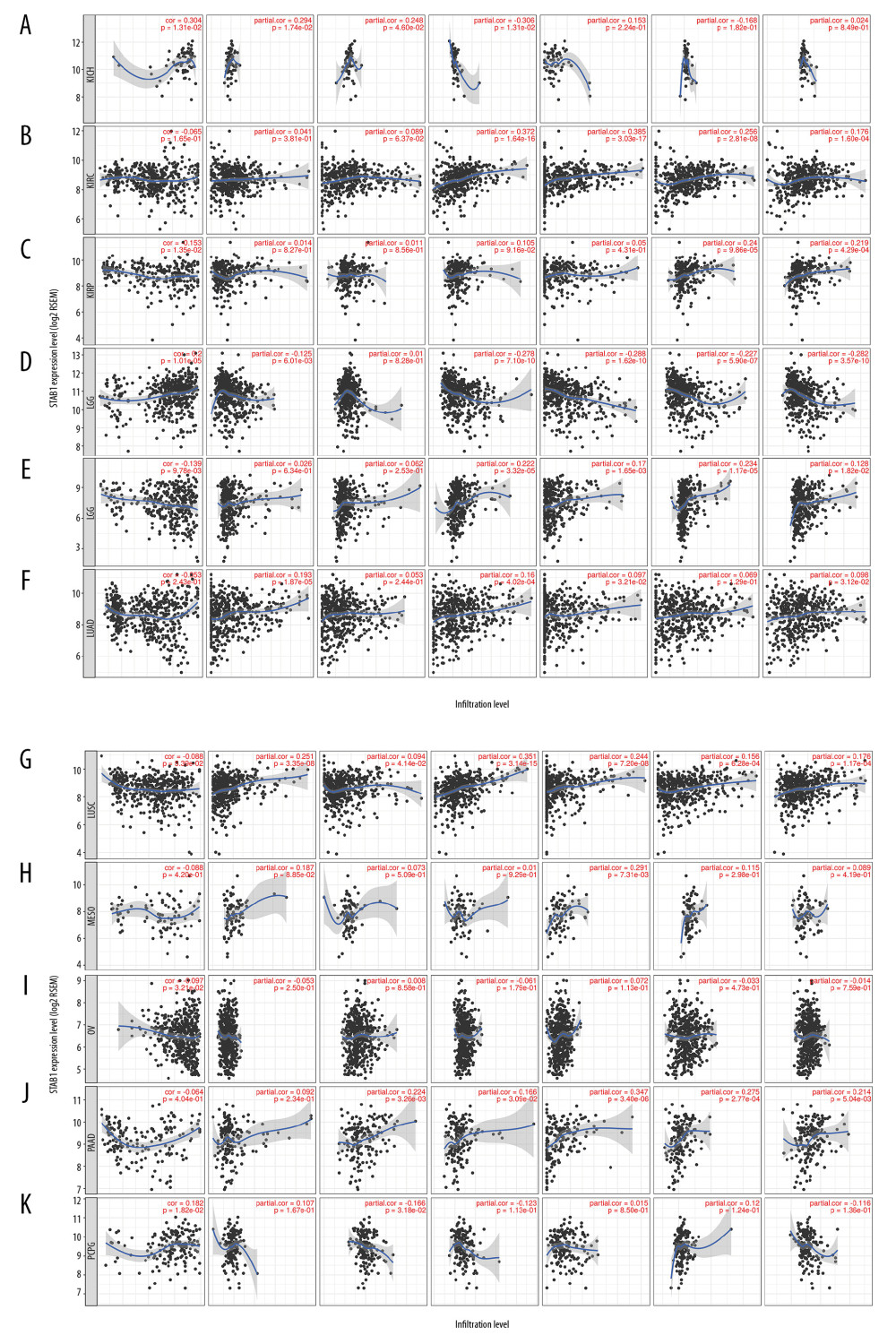 Figure 7. Association of SATB1 expression with immune infiltration levels in (A) LGG. (B) LIHC. (C) LUAD. (D) LUSC. (E) MESO. (F) OV. (G) PAAD. (H) PCPG. (I) READ. (J) SARC. (K) SKCM. LGG – low-grade gliomas; LIHC – liver hepatocellular carcinoma; LUAD – lung adenocarcinoma; LUSC – lung squamous cell carcinoma; MESO – mesothelioma; OV – ovarian; PAAD – pancreatic adenocarcinoma; PCPG – paraganglioma; READ – rectum adenocarcinoma; SARC – sarcoma; SKCM – skin cutaneous melanoma.
Figure 7. Association of SATB1 expression with immune infiltration levels in (A) LGG. (B) LIHC. (C) LUAD. (D) LUSC. (E) MESO. (F) OV. (G) PAAD. (H) PCPG. (I) READ. (J) SARC. (K) SKCM. LGG – low-grade gliomas; LIHC – liver hepatocellular carcinoma; LUAD – lung adenocarcinoma; LUSC – lung squamous cell carcinoma; MESO – mesothelioma; OV – ovarian; PAAD – pancreatic adenocarcinoma; PCPG – paraganglioma; READ – rectum adenocarcinoma; SARC – sarcoma; SKCM – skin cutaneous melanoma. 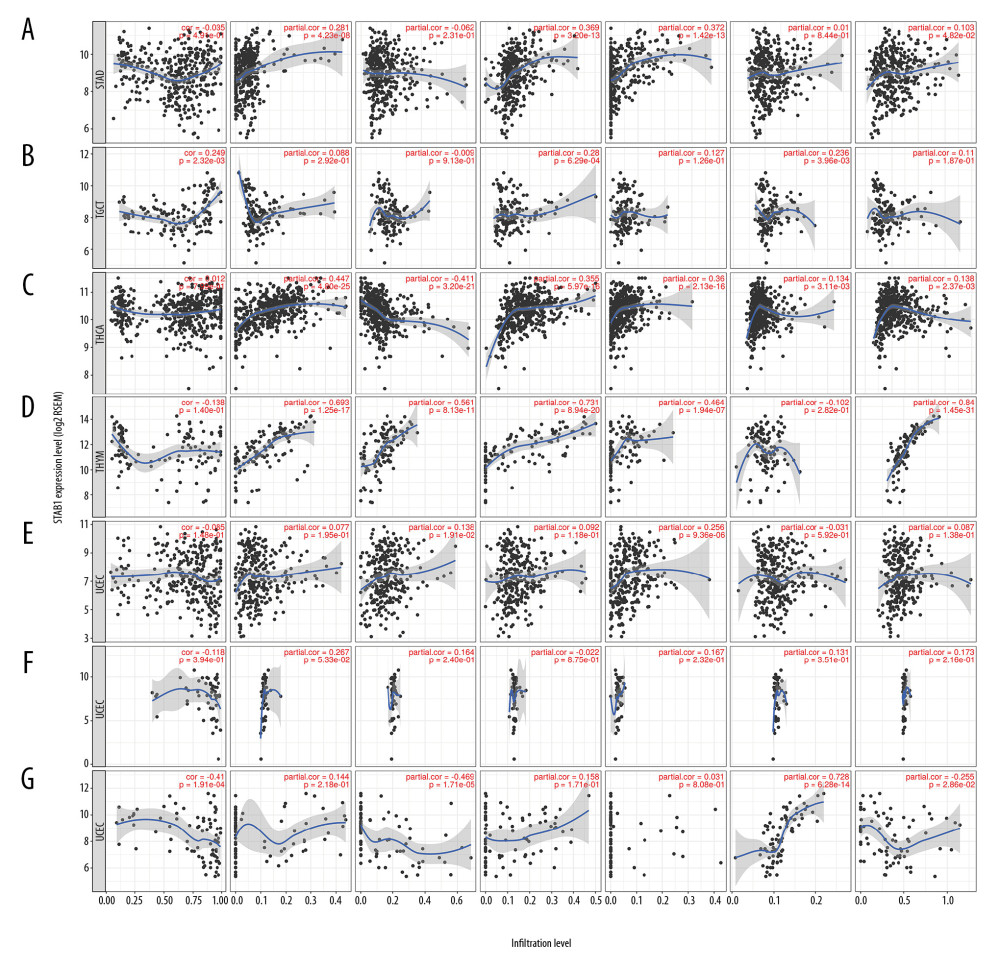 Figure 8. Association of SATB1 expression with immune infiltration levels in (A) STAD. (B) TGCT. (C) THCA. (D) THYM. (E) UCEC. (F) UCS. (G) UVM. STAD – stomach adenocarcinoma; TGCT – testicular germ cell tumors; THCA – thyroid carcinoma; THYM – thymoma; UCEC – uterine corpus endometrial carcinoma; UCS – uterine carcinosarcoma. UVM – uveal melanoma.
Figure 8. Association of SATB1 expression with immune infiltration levels in (A) STAD. (B) TGCT. (C) THCA. (D) THYM. (E) UCEC. (F) UCS. (G) UVM. STAD – stomach adenocarcinoma; TGCT – testicular germ cell tumors; THCA – thyroid carcinoma; THYM – thymoma; UCEC – uterine corpus endometrial carcinoma; UCS – uterine carcinosarcoma. UVM – uveal melanoma. 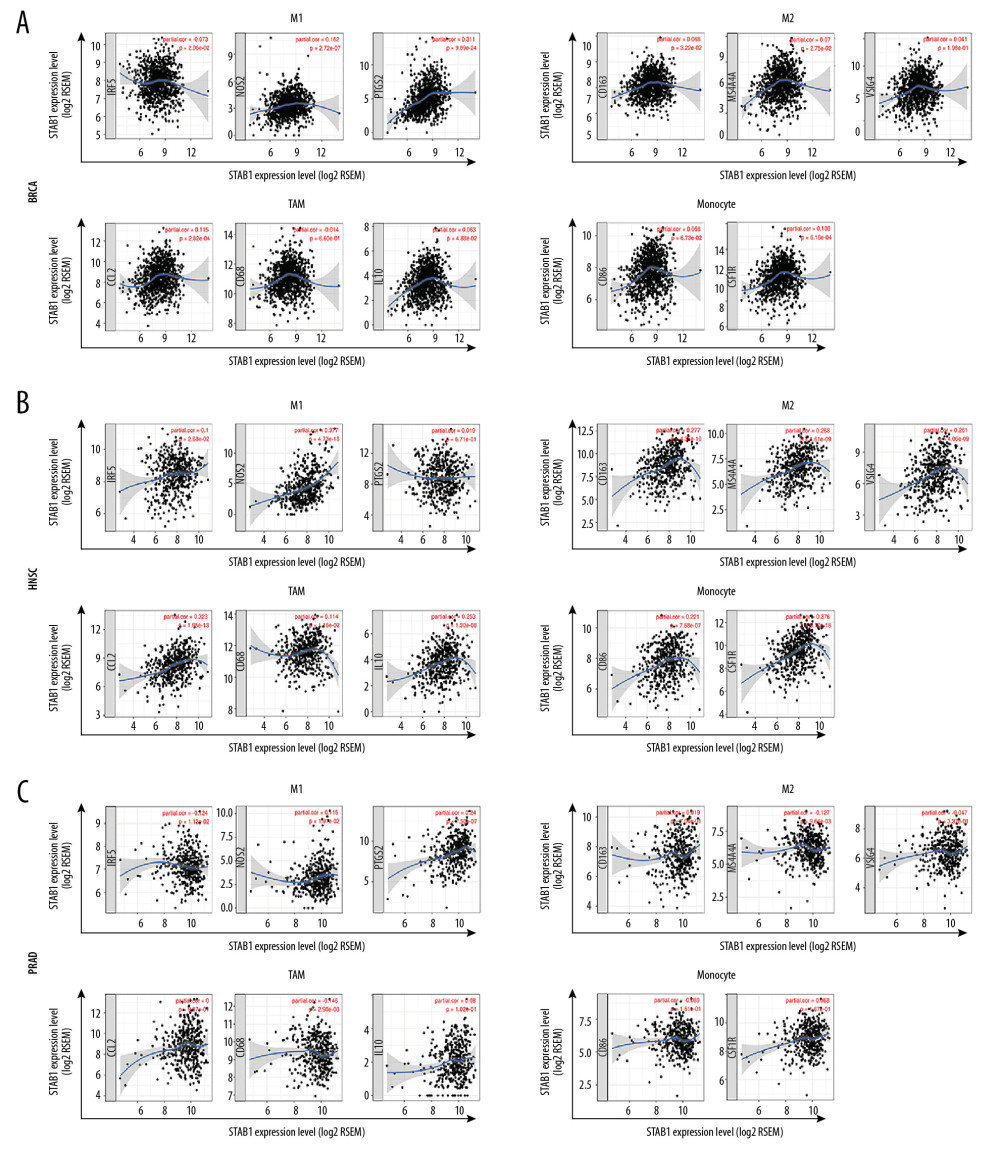 Figure 9. SATB1 expression correlated with macrophage polarization in BRAC, HNSC, and PRAD. Markers include CD86 and CSF1R of monocytes; CCL2, CD68, and IL10 of TAMs; NOS2, IRF5, and PTGS2 of M1 macrophages; and CD163, VSIG4, and MS4A4A of M2 macrophages. (A) Scatterplots of correlations between SATB1 expression and gene markers of M1 macrophages, M2 macrophages, TAMs, and monocytes in BRAC (n=1093). (B) Scatterplots of correlations between SATB1 expression and gene markers of M1 macrophages, M2 macrophages, TAMs, and monocytes in HNSC (n=520). (C) Scatterplots of correlations between SATB1 expression and gene markers of M1 macrophages, M2 macrophages, TAMs, and monocytes in PRAD (n=497). BRCA – breast invasive carcinoma; HNSC – head and neck cancer; PRAD – prostate adenocarcinoma; TAMs – tumor associated macrophages.
Figure 9. SATB1 expression correlated with macrophage polarization in BRAC, HNSC, and PRAD. Markers include CD86 and CSF1R of monocytes; CCL2, CD68, and IL10 of TAMs; NOS2, IRF5, and PTGS2 of M1 macrophages; and CD163, VSIG4, and MS4A4A of M2 macrophages. (A) Scatterplots of correlations between SATB1 expression and gene markers of M1 macrophages, M2 macrophages, TAMs, and monocytes in BRAC (n=1093). (B) Scatterplots of correlations between SATB1 expression and gene markers of M1 macrophages, M2 macrophages, TAMs, and monocytes in HNSC (n=520). (C) Scatterplots of correlations between SATB1 expression and gene markers of M1 macrophages, M2 macrophages, TAMs, and monocytes in PRAD (n=497). BRCA – breast invasive carcinoma; HNSC – head and neck cancer; PRAD – prostate adenocarcinoma; TAMs – tumor associated macrophages. Tables
Table 1. Relation between SATB1 expression and patient prognosis in different cancer using PrognosScan database.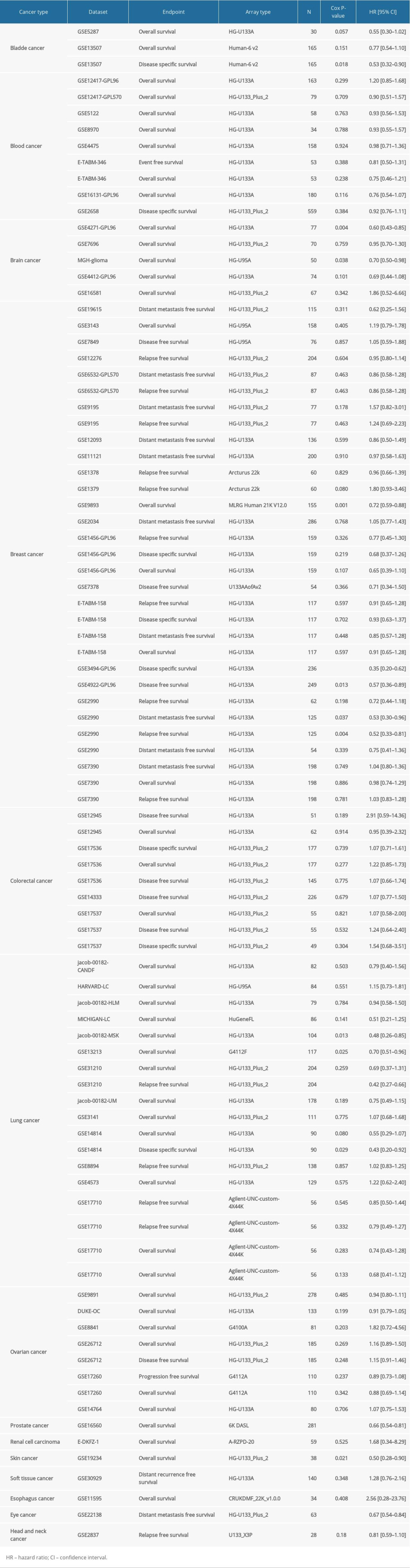 Table 2. Correlation of SATB1 expression with prognostic values in diverse types of cancer in GEPIA.
Table 2. Correlation of SATB1 expression with prognostic values in diverse types of cancer in GEPIA.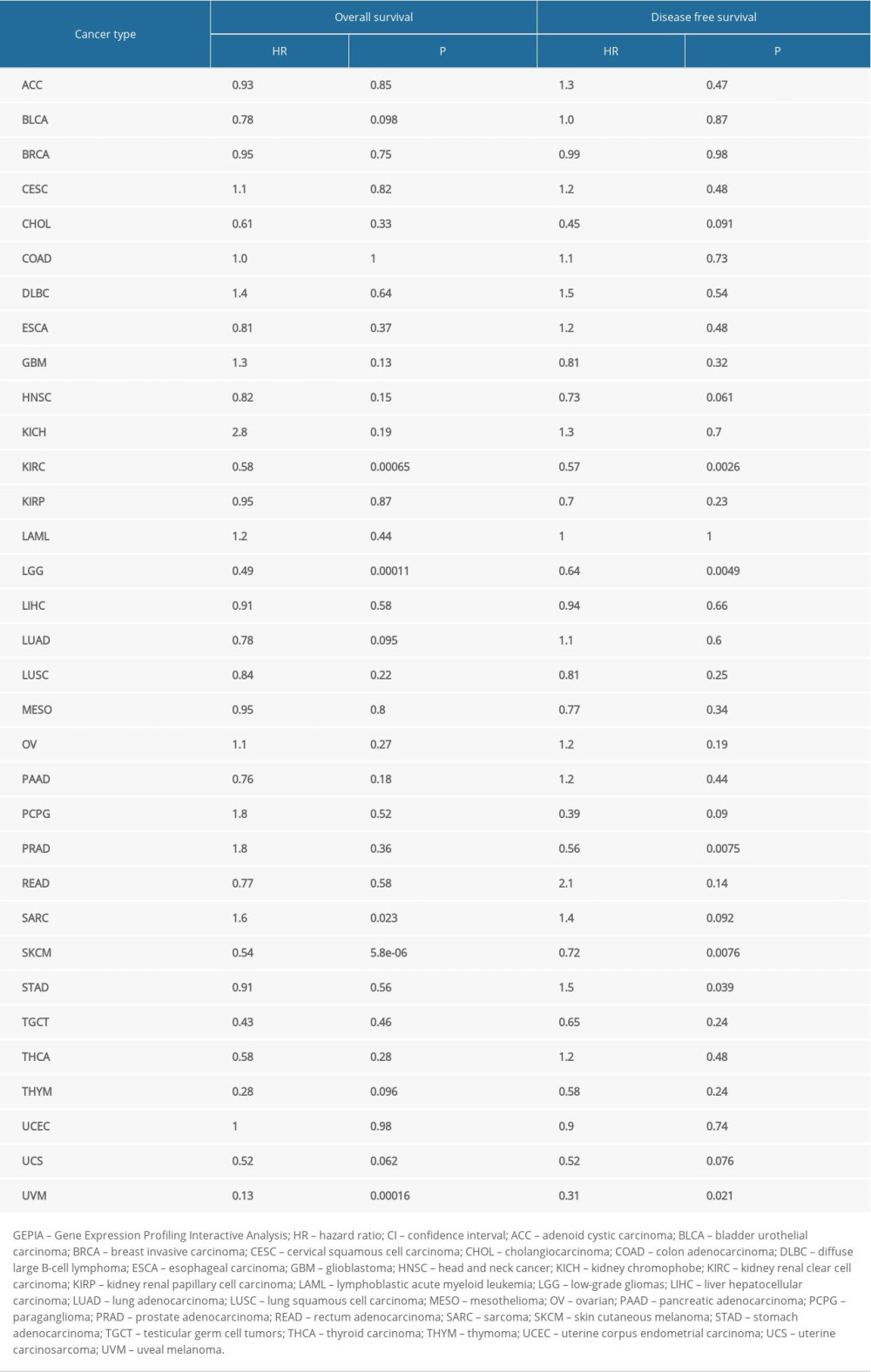 Table 3. Correlation of SATB1 mRNA expression and prognosis in breast cancer with different clinicopathological characters by Kaplan-Meier plotter.
Table 3. Correlation of SATB1 mRNA expression and prognosis in breast cancer with different clinicopathological characters by Kaplan-Meier plotter.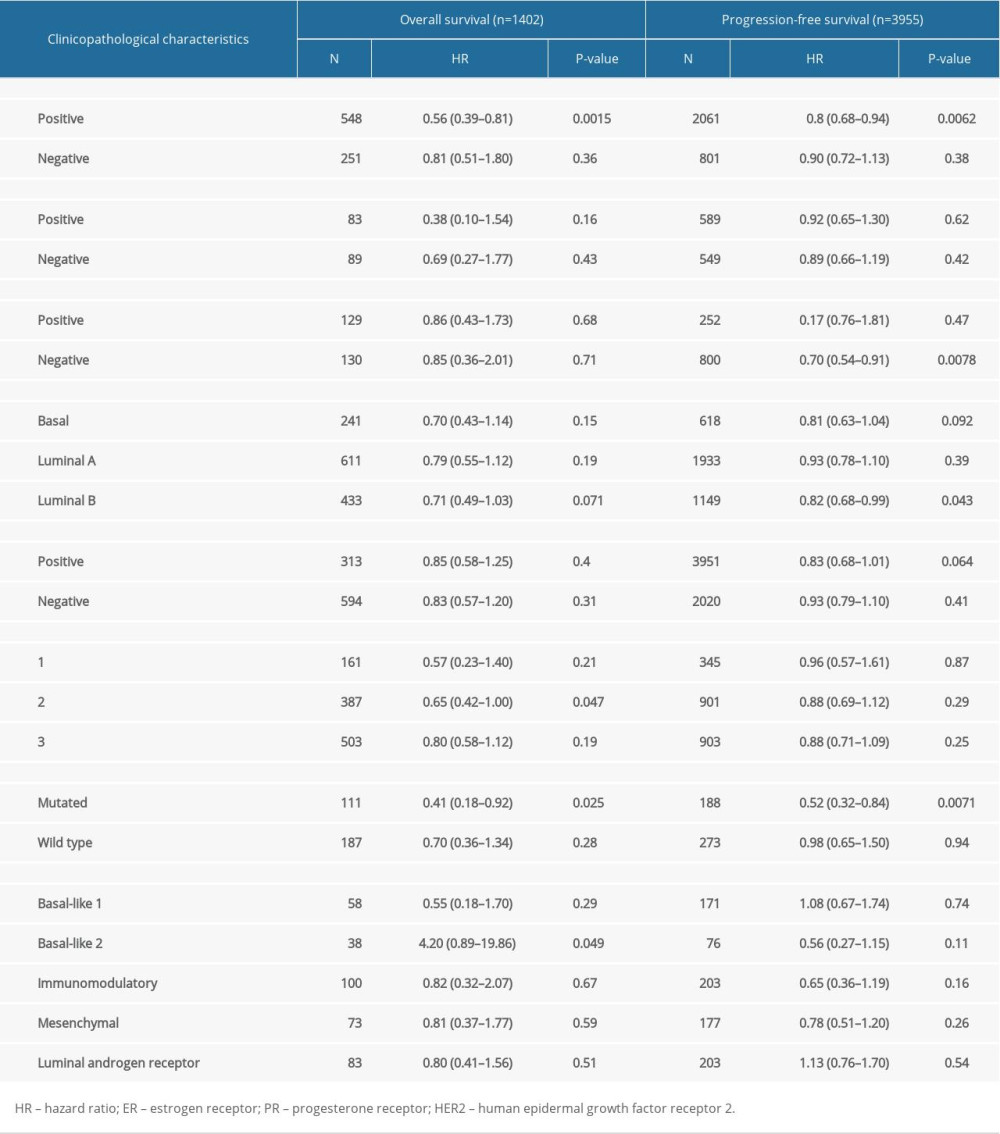 Table 4. Correlation analysis between SATB1 and relate genes and markers of immune cells in TIMER.
Table 4. Correlation analysis between SATB1 and relate genes and markers of immune cells in TIMER.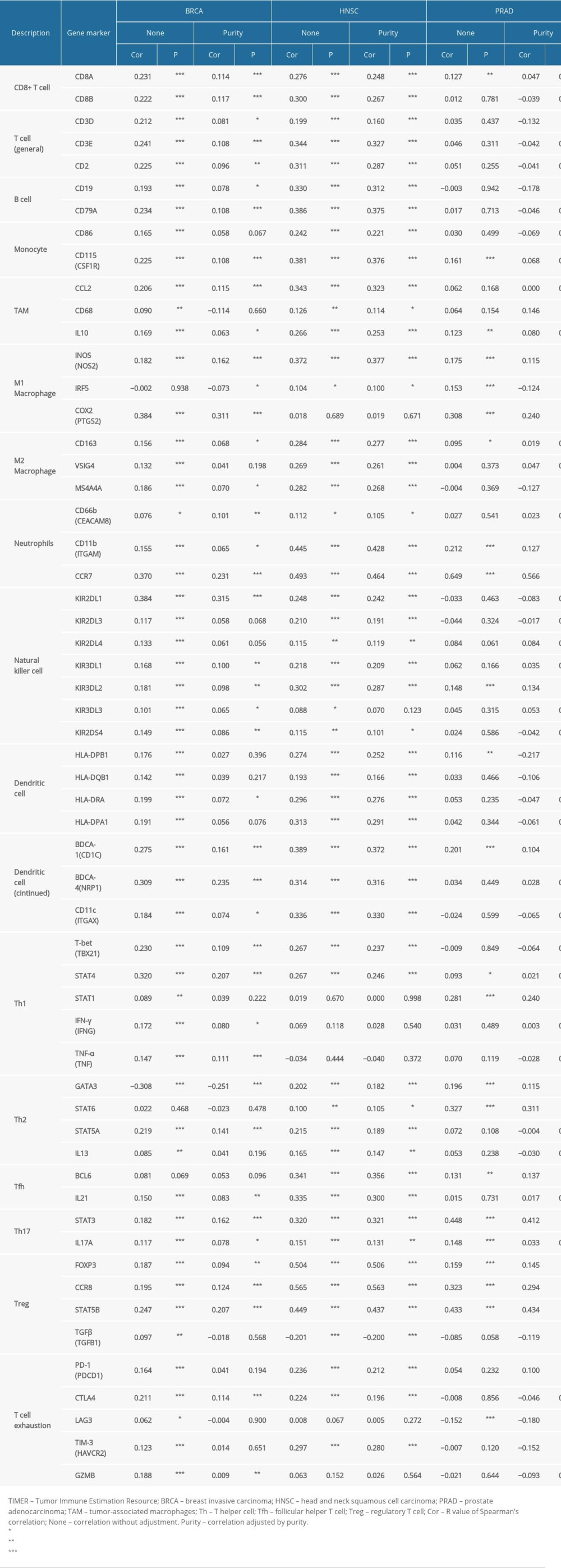
References
1. Bray F, Ferlay J, Soerjomataram I, Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries: Cancer J Clin, 2018; 68; 394-424
2. Esteva FJ, Hubbard-Lucey VM, Tang J, Pusztai L, Immunotherapy and targeted therapy combinations in metastatic breast cancer: Lancet Oncol, 2019; 20; e175-86
3. Sim F, Leidner R, Bell RB, Immunotherapy for head and neck cancer: Hematol Oncol Clin North Am, 2019; 33; 301-21
4. Henegan JC, Sonpavde G, Promising immunotherapy for prostate cancer: Expert Opin Biol Ther, 2018; 18; 109-20
5. Esteva FJ, Wang J, Lin F, CD40 signaling predicts response to preoperative trastuzumab and concomitant paclitaxel followed by 5-fluorouracil, epirubicin, and cyclophosphamide in HER-2-overexpressing breast cancer: Breast Cancer Res, 2007; 9; R87
6. Przybylski K, Majchrzak E, Weselik L, Golusinski W, Immunotherapy of head and neck squamous cell carcinoma (HNSCC). Immune checkpoint blockade: Otolaryngol Pol, 2018; 72; 10-16
7. Seiwert TY, Burtness B, Mehra R, Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): An open-label, multicentre, phase 1b trial: Lancet Oncol, 2016; 17; 956-65
8. Schepisi G, Farolfi A, Conteduca V, Immunotherapy for prostate cancer: Where we are headed: Int J Mol Sci, 2017; 18 pii: E2627
9. Salmi M, Macrophages and cancer: Duodecim, 2017; 133; 829-37
10. Treffers LW, Hiemstra IH, Kuijpers TW, Neutrophils in cancer: Immunol Rev, 2016; 273; 312-28
11. Dickinson LA, Joh T, Kohwi Y, Kohwi-Shigematsu T, A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition: Cell, 1992; 70; 631-45
12. Purbey PK, Singh S, Kumar PP, PDZ domain-mediated dimerization and homeodomain-directed specificity are required for high-affinity DNA binding by SATB1: Nucleic Acids Res, 2008; 36; 2107-22
13. Ding M, Pan J, Guo Z, SATB1 is a novel molecular target for cancer therapy: Cancer Invest, 2018; 36; 28-36
14. Alvarez JD, Yasui DH, Niida H, The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development: Genes Dev, 2000; 14; 521-35
15. Stephen TL, Payne KK, Chaurio RA, SATB1 expression governs epigenetic repression of PD-1 in tumor-reactive T cells: Immunity, 2017; 46; 51-64
16. Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles: Neoplasia, 2007; 9; 166-80
17. Li T, Fan J, Wang B, TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells: Cancer Res, 2017; 77; e108-10
18. Mizuno H, Kitada K, Nakai K, Sarai A, PrognoScan: A new database for meta-analysis of the prognostic value of genes: BMC Med Genomics, 2009; 2; 18
19. Nagy A, Lanczky A, Menyhart O, Gyorffy B, Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets: Sci Rep, 2018; 8; 9227
20. Aran D, Sirota M, Butte AJ, Systematic pan-cancer analysis of tumour purity: Nat Commun, 2015; 6; 8971
21. Pan JH, Zhou H, Cooper L, LAYN is a prognostic biomarker and correlated with immune infiltrates in gastric and colon cancers: Front Immunol, 2019; 10; 6
22. Sotiriou C, Wirapati P, Loi S, Gene expression profiling in breast cancer: Understanding the molecular basis of histologic grade to improve prognosis: J Natl Cancer Inst, 2006; 98; 262-72
23. Lovitch SB, Rodig SJ, The role of surgical pathology in guiding cancer immunotherapy: Annu Rev Pathol, 2016; 11; 313-41
24. Sunkara KP, Gupta G, Hansbro PM, Functional relevance of SATB1 in immune regulation and tumorigenesis: Biomed Pharmacother, 2018; 104; 87-93
25. Mir R, Pradhan SJ, Patil P, Wnt/beta-catenin signaling regulated SATB1 promotes colorectal cancer tumorigenesis and progression: Oncogene, 2016; 35; 1679-91
26. Pan Z, Jing W, He K, SATB1 is correlated with progression and metastasis of breast cancers: A meta-analysis: Cell Physiol Biochem, 2016; 38; 1975-83
27. Zhai S, Xue J, Wang Z, Hu L, High expression of special AT-rich sequence binding protein-1 predicts esophageal squamous cell carcinoma relapse and poor prognosis: Oncol Lett, 2017; 14; 7455-60
28. Tesone AJ, Rutkowski MR, Brencicova E, SATB1 overexpression drives tumor-promoting activities in cancer-associated dendritic cells: Cell Rep, 2016; 14; 1774-86
29. Cubillos-Ruiz JR, Baird JR, Tesone AJ: Cancer Res, 2012; 72; 1683-93
30. Fredholm S, Willerslev-Olsen A, Met O, SATB1 in malignant T cells: J Invest Dermatol, 2018; 138; 1805-15
31. Kondo M, Tanaka Y, Kuwabara T, SATB1 plays a critical role in establishment of immune tolerance: J Immunol, 2016; 196; 563-72
Figures
 Figure 1. Flow chart of the study procedures.
Figure 1. Flow chart of the study procedures. Figure 2. The expression of SATB1 in human cancers. (A) High or low SATB1 expression in data sets of different cancers in Oncomine database. (B) SATB1 expression levels in different tumor types from Tumor Immune Estimation Resource database (* P<0.05; ** P<0.01; *** P<0.001).
Figure 2. The expression of SATB1 in human cancers. (A) High or low SATB1 expression in data sets of different cancers in Oncomine database. (B) SATB1 expression levels in different tumor types from Tumor Immune Estimation Resource database (* P<0.05; ** P<0.01; *** P<0.001).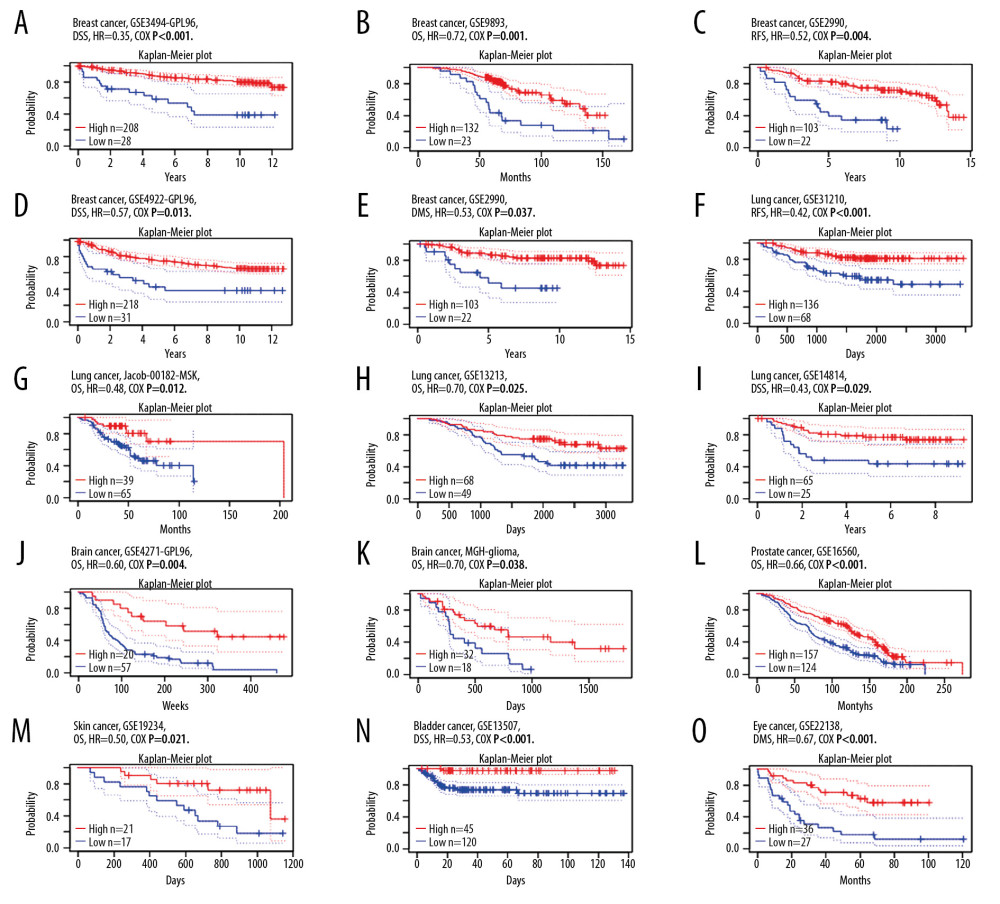 Figure 3. Kaplan-Meier survival curves comparing the increased and decreased expression of SATB1 in different types of cancer in PrognoScan. (A–E) Survival curves of DSS, OS, RFS, DFS, and DMFS in breast cancer cohorts [GSE3494-GPL96 (n=236), GSE9893 (n=155), GSE2990 (n=125), and GSE4922 (n=249)]. (F–I) Survival curves of RFS, OS, and DSS in lung cancer cohorts [GSE31210 (n=204), Jacob-00182-MSK (n=104), GSE13213 (n=117), and GSE14814 (n=90)]. (J, K) Survival curves of OS in brain cancer cohorts [GSE4271-GPL96 (n=77), MGH-glioma (n=50)]. (L) Survival curves of OS in prostate cancer cohorts [GSE16560 (n=281)]. (M) Survival curves of OS in skin cancer cohorts [GSE19234 (n=38)]. (N) Survival curves of DSS in bladder cancer cohorts [GSE13507 (n=165)]. (O) Survival curves of DSS in eye cancer cohorts [GSE22138 (n=63)]. DSS – disease-specific survival; OS – overall survival; RFS – relapse-free survival; DFS – disease-free survival; DMFS – distant metastasis free survival.
Figure 3. Kaplan-Meier survival curves comparing the increased and decreased expression of SATB1 in different types of cancer in PrognoScan. (A–E) Survival curves of DSS, OS, RFS, DFS, and DMFS in breast cancer cohorts [GSE3494-GPL96 (n=236), GSE9893 (n=155), GSE2990 (n=125), and GSE4922 (n=249)]. (F–I) Survival curves of RFS, OS, and DSS in lung cancer cohorts [GSE31210 (n=204), Jacob-00182-MSK (n=104), GSE13213 (n=117), and GSE14814 (n=90)]. (J, K) Survival curves of OS in brain cancer cohorts [GSE4271-GPL96 (n=77), MGH-glioma (n=50)]. (L) Survival curves of OS in prostate cancer cohorts [GSE16560 (n=281)]. (M) Survival curves of OS in skin cancer cohorts [GSE19234 (n=38)]. (N) Survival curves of DSS in bladder cancer cohorts [GSE13507 (n=165)]. (O) Survival curves of DSS in eye cancer cohorts [GSE22138 (n=63)]. DSS – disease-specific survival; OS – overall survival; RFS – relapse-free survival; DFS – disease-free survival; DMFS – distant metastasis free survival. Figure 4. Kaplan-Meier survival curves comparing the expression of SATB1 in different types of cancer in Kaplan-Meier plotter database. (A, B) OS and RFS of breast cancer (n=1402, n=977). (C, D) OS and PFS of lung cancer (n=1145, n=982). (E, F) OS and PFS of gastric cancer (n 631, n=522). (G, H) OS and PFS of ovarian cancer (n=655, n=1435). OS – overall survival; RFS – relapse-free survival; PFS – progression-free survival.
Figure 4. Kaplan-Meier survival curves comparing the expression of SATB1 in different types of cancer in Kaplan-Meier plotter database. (A, B) OS and RFS of breast cancer (n=1402, n=977). (C, D) OS and PFS of lung cancer (n=1145, n=982). (E, F) OS and PFS of gastric cancer (n 631, n=522). (G, H) OS and PFS of ovarian cancer (n=655, n=1435). OS – overall survival; RFS – relapse-free survival; PFS – progression-free survival. Figure 5. SATB1 expression is negatively correlated with tumor purity and positively correlated with infiltration levels of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells in BRCA, HNSC, and PRAD. (A) Correlation of SATB1 expression with immune infiltration level in BRAC (n=1093). (B) Correlation of SATB1 expression with immune infiltration level in HNSC (n=520). (C) Correlation of SATB1 expression with immune infiltration level in PRAD (n=497). BRCA – breast invasive carcinoma; HNSC – head and neck cancer; PRAD – prostate adenocarcinoma.
Figure 5. SATB1 expression is negatively correlated with tumor purity and positively correlated with infiltration levels of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells in BRCA, HNSC, and PRAD. (A) Correlation of SATB1 expression with immune infiltration level in BRAC (n=1093). (B) Correlation of SATB1 expression with immune infiltration level in HNSC (n=520). (C) Correlation of SATB1 expression with immune infiltration level in PRAD (n=497). BRCA – breast invasive carcinoma; HNSC – head and neck cancer; PRAD – prostate adenocarcinoma. Figure 6. Association of SATB1 expression with immune infiltration levels in (A) ACC. (B) BLCA. (C) CESC. (D) CHOL. (E) COAD. (F) DLBC. (G) ESCA. (H) GBM. (I) KICH. (J) KIRC. (K) KIRP. ACC – adenoid cystic carcinoma; BLCA – bladder urothelial carcinoma; CESC – cervical squamous cell carcinoma; CHOL – cholangiocarcinoma; COAD – colon adenocarcinoma; DLBC – diffuse large B-cell lymphoma; ESCA – esophageal carcinoma; GBM – glioblastoma; KICH – kidney chromophobe; KIRC – kidney renal clear cell carcinoma; KIRP – kidney renal papillary cell carcinoma.
Figure 6. Association of SATB1 expression with immune infiltration levels in (A) ACC. (B) BLCA. (C) CESC. (D) CHOL. (E) COAD. (F) DLBC. (G) ESCA. (H) GBM. (I) KICH. (J) KIRC. (K) KIRP. ACC – adenoid cystic carcinoma; BLCA – bladder urothelial carcinoma; CESC – cervical squamous cell carcinoma; CHOL – cholangiocarcinoma; COAD – colon adenocarcinoma; DLBC – diffuse large B-cell lymphoma; ESCA – esophageal carcinoma; GBM – glioblastoma; KICH – kidney chromophobe; KIRC – kidney renal clear cell carcinoma; KIRP – kidney renal papillary cell carcinoma. Figure 7. Association of SATB1 expression with immune infiltration levels in (A) LGG. (B) LIHC. (C) LUAD. (D) LUSC. (E) MESO. (F) OV. (G) PAAD. (H) PCPG. (I) READ. (J) SARC. (K) SKCM. LGG – low-grade gliomas; LIHC – liver hepatocellular carcinoma; LUAD – lung adenocarcinoma; LUSC – lung squamous cell carcinoma; MESO – mesothelioma; OV – ovarian; PAAD – pancreatic adenocarcinoma; PCPG – paraganglioma; READ – rectum adenocarcinoma; SARC – sarcoma; SKCM – skin cutaneous melanoma.
Figure 7. Association of SATB1 expression with immune infiltration levels in (A) LGG. (B) LIHC. (C) LUAD. (D) LUSC. (E) MESO. (F) OV. (G) PAAD. (H) PCPG. (I) READ. (J) SARC. (K) SKCM. LGG – low-grade gliomas; LIHC – liver hepatocellular carcinoma; LUAD – lung adenocarcinoma; LUSC – lung squamous cell carcinoma; MESO – mesothelioma; OV – ovarian; PAAD – pancreatic adenocarcinoma; PCPG – paraganglioma; READ – rectum adenocarcinoma; SARC – sarcoma; SKCM – skin cutaneous melanoma. Figure 8. Association of SATB1 expression with immune infiltration levels in (A) STAD. (B) TGCT. (C) THCA. (D) THYM. (E) UCEC. (F) UCS. (G) UVM. STAD – stomach adenocarcinoma; TGCT – testicular germ cell tumors; THCA – thyroid carcinoma; THYM – thymoma; UCEC – uterine corpus endometrial carcinoma; UCS – uterine carcinosarcoma. UVM – uveal melanoma.
Figure 8. Association of SATB1 expression with immune infiltration levels in (A) STAD. (B) TGCT. (C) THCA. (D) THYM. (E) UCEC. (F) UCS. (G) UVM. STAD – stomach adenocarcinoma; TGCT – testicular germ cell tumors; THCA – thyroid carcinoma; THYM – thymoma; UCEC – uterine corpus endometrial carcinoma; UCS – uterine carcinosarcoma. UVM – uveal melanoma. Figure 9. SATB1 expression correlated with macrophage polarization in BRAC, HNSC, and PRAD. Markers include CD86 and CSF1R of monocytes; CCL2, CD68, and IL10 of TAMs; NOS2, IRF5, and PTGS2 of M1 macrophages; and CD163, VSIG4, and MS4A4A of M2 macrophages. (A) Scatterplots of correlations between SATB1 expression and gene markers of M1 macrophages, M2 macrophages, TAMs, and monocytes in BRAC (n=1093). (B) Scatterplots of correlations between SATB1 expression and gene markers of M1 macrophages, M2 macrophages, TAMs, and monocytes in HNSC (n=520). (C) Scatterplots of correlations between SATB1 expression and gene markers of M1 macrophages, M2 macrophages, TAMs, and monocytes in PRAD (n=497). BRCA – breast invasive carcinoma; HNSC – head and neck cancer; PRAD – prostate adenocarcinoma; TAMs – tumor associated macrophages.
Figure 9. SATB1 expression correlated with macrophage polarization in BRAC, HNSC, and PRAD. Markers include CD86 and CSF1R of monocytes; CCL2, CD68, and IL10 of TAMs; NOS2, IRF5, and PTGS2 of M1 macrophages; and CD163, VSIG4, and MS4A4A of M2 macrophages. (A) Scatterplots of correlations between SATB1 expression and gene markers of M1 macrophages, M2 macrophages, TAMs, and monocytes in BRAC (n=1093). (B) Scatterplots of correlations between SATB1 expression and gene markers of M1 macrophages, M2 macrophages, TAMs, and monocytes in HNSC (n=520). (C) Scatterplots of correlations between SATB1 expression and gene markers of M1 macrophages, M2 macrophages, TAMs, and monocytes in PRAD (n=497). BRCA – breast invasive carcinoma; HNSC – head and neck cancer; PRAD – prostate adenocarcinoma; TAMs – tumor associated macrophages. Tables
 Table 1. Relation between SATB1 expression and patient prognosis in different cancer using PrognosScan database.
Table 1. Relation between SATB1 expression and patient prognosis in different cancer using PrognosScan database. Table 2. Correlation of SATB1 expression with prognostic values in diverse types of cancer in GEPIA.
Table 2. Correlation of SATB1 expression with prognostic values in diverse types of cancer in GEPIA. Table 3. Correlation of SATB1 mRNA expression and prognosis in breast cancer with different clinicopathological characters by Kaplan-Meier plotter.
Table 3. Correlation of SATB1 mRNA expression and prognosis in breast cancer with different clinicopathological characters by Kaplan-Meier plotter. Table 4. Correlation analysis between SATB1 and relate genes and markers of immune cells in TIMER.
Table 4. Correlation analysis between SATB1 and relate genes and markers of immune cells in TIMER. Table 1. Relation between SATB1 expression and patient prognosis in different cancer using PrognosScan database.
Table 1. Relation between SATB1 expression and patient prognosis in different cancer using PrognosScan database. Table 2. Correlation of SATB1 expression with prognostic values in diverse types of cancer in GEPIA.
Table 2. Correlation of SATB1 expression with prognostic values in diverse types of cancer in GEPIA. Table 3. Correlation of SATB1 mRNA expression and prognosis in breast cancer with different clinicopathological characters by Kaplan-Meier plotter.
Table 3. Correlation of SATB1 mRNA expression and prognosis in breast cancer with different clinicopathological characters by Kaplan-Meier plotter. Table 4. Correlation analysis between SATB1 and relate genes and markers of immune cells in TIMER.
Table 4. Correlation analysis between SATB1 and relate genes and markers of immune cells in TIMER. In Press
07 Mar 2024 : Clinical Research
Knowledge of and Attitudes Toward Clinical Trials: A Questionnaire-Based Study of 179 Male Third- and Fourt...Med Sci Monit In Press; DOI: 10.12659/MSM.943468
08 Mar 2024 : Animal Research
Modification of Experimental Model of Necrotizing Enterocolitis (NEC) in Rat Pups by Single Exposure to Hyp...Med Sci Monit In Press; DOI: 10.12659/MSM.943443
18 Apr 2024 : Clinical Research
Comparative Analysis of Open and Closed Sphincterotomy for the Treatment of Chronic Anal Fissure: Safety an...Med Sci Monit In Press; DOI: 10.12659/MSM.944127
08 Mar 2024 : Laboratory Research
Evaluation of Retentive Strength of 50 Endodontically-Treated Single-Rooted Mandibular Second Premolars Res...Med Sci Monit In Press; DOI: 10.12659/MSM.944110
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952








