14 July 2020: Clinical Research
Long Non-Coding RNA THOR Enhances the Stem Cell-Like Traits of Triple-Negative Breast Cancer Cells Through Activating β-Catenin Signaling
Binbin Wang1ABCE, Qiang Ye2BCD, Chuantao Zou1AEG*DOI: 10.12659/MSM.923507
Med Sci Monit 2020; 26:e923507
Abstract
BACKGROUND: The oncogenic roles of lncRNA THOR have been revealed in several tumors, however, its functions in breast cancer are still unclear.
MATERIAL AND METHODS: Real-time quantitative polymerase chain reaction (RT-qPCR) was used to detect THOR expression in clinical samples and the expression of stemness regulatory factors. ALDH1 assay and sphere-formation analysis were constructed to examine the stemness of cells. Cell viability assay was constructed to determine the cell proliferation capacity. In vitro RNA-RNA interaction and messenger RNA (mRNA) stability assays were performed to explore the mechanisms.
RESULTS: THOR was overexpressed in triple-negative breast cancer (TNBC) compared to that in luminal A- and B-type breast cancer. THOR silencing reduced TNBC cell stemness, which was evident by the decreased sphere-formation ability, stemness marker expression and ALDH1 activity. Mechanistically, THOR directly bound to β-catenin mRNA, enhanced β-catenin mRNA stability and thus increased its expression. Furthermore, overexpression of β-catenin partially diminished THOR silencing-mediated inhibition on TNBC cell stemness.
CONCLUSIONS: This work proposes that THOR facilitates TNBC cell stemness through activating β-catenin signaling.
Keywords: Neoplastic Stem Cells, Triple Negative Breast Neoplasms, RNA, Messenger, beta Catenin
Background
The incidence of breast cancer, which can be divided into luminal A, luminal B and triple-negative breast cancer (TNBC) sub-types, ranks the first in female malignant tumors [1]. The cell origin, gene mutation, metastasis potential, disease progression, therapeutic response, and clinical outcome of each subtype are substantially different [2]. TNBC refers to breast cancer with negative estrogen, negative progesterone and negative human epidermal growth factor receptor, and exhibits the worst prognosis [2]. TNBC is prone to recurrence and metastasis, and lacks targeted treatment. Cancer stem cells (CSCs), which contribute to cancer cell progression, play critical effects in recurrence, metastasis, and drug resistance [3]. The proportion of CSCs in TNBC is significantly higher than other subtypes and plenty of evidences indicates that CSCs in TNBC is considered to be a lead factor in its poor prognosis [3]. Thus, targeting CSCs is expected to be an effective means to improve the poor prognosis of TNBC.
LncRNA (long non-coding RNA) is a kind of regulatory non-coding RNA [4]. Its transcript length is more than 200 nt, and it has no obvious open reading frame (ORF) without protein-coding ability [4]. Recently, a large number of studies have shown that lncRNA can modulate gene expression at the transcriptional or post-transcriptional level through binding to DNA, RNA, or protein [4]. With the rapid development of high-throughput sequencing and chip-detection technologies, many kinds of lncRNAs with abnormal expression have been found in breast cancer [5]. Some lncRNAs have been identified to be the important regulators of breast cancer occurrence and development and can be used as effective and accurate indicators for breast cancer diagnosis, for instance, HOTAIR [6], MALAT1 [7], LSINCT5 [8], H19 [4], and BC200 [9], while lncRNA XIST [10] and GAS5 [11] are lowly expressed in breast cancer. HOTAIR expression is positively associated with the poor prognosis of breast cancer [6], and so is MALAT1 expression for breast cancer metastasis [7]. In addition, the abnormal expression of some lncRNAs is closely related to breast cancer occurrence and development. LncRNA HOTAIR promotes breast cancer metastasis of by inhibiting the binding of BRCA1 to EZH2 [12]. H19 increases the proliferation of breast cancer cells by regulating cell cycle through inducing E2F1 and c-Myc expression [13]. LncRNA-NKILA can directly bind to NF-kappa b/I-kappa B, reduce the transcriptional activity of NF-kappa B and thus suppress breast cancer metastasis [14]. LncRNA THOR was initially identified as an ultra-conserved lncRNA in 2017, exhibits an exclusive expression in testis and contributes to tumor growth [15]. Further studies have shown that THOR promotes cell survival and proliferation of osteosarcoma [16], renal cell carcinoma [17], and hepatocellular carcinoma [18]. Notably, recent works showed that THOR enhances the CSC-like traits of gastric cancer cells [19], osteosarcoma cells [20], and liver cancer cells [21]. However, the effects and underlying mechanisms of THOR in breast cancer, especially in TNBC, are unclear.
In our study, THOR expression was determined in luminal A and luminal B breast cancer and TNBC tissues. It was found that THOR was highly expressed in TNBC cells and tissues. Additionally, THOR knockdown reduced TNBC cell stemness through detecting the expression of CSC regulatory factors, ALDH1 activity and sphere-formation ability. Further mechanistic studies revealed that THOR directly interacted with β-catenin messenger RNA (mRNA), enhanced β-catenin mRNA stability and thus increased its expression, which is essential for THOR-induced effects on TNBC cell stemness.
Material and Methods
CLINICAL SAMPLES:
Forty-three pairs of breast tumors (including 15 TNBC sub-types, 17 luminal A sub-types and 11 luminal B sub-types) with adjacent mammary epithelial tissues were obtained from 43 patients who underwent surgery at the Suizhou Hospital Affiliated to Hubei University of Medicine from February 2017 to January 2019 and all cases did not metastasize. Written informed consent has been obtained from each patient. Approval from the Suizhou Hospital Affiliated to Hubei University of Medicine Ethics Committee was obtained for the use of these clinical materials for research purposes. All experiments conform to the Declaration of Helsinki.
CELL CULTURE:
Human TNBC lines MDA-MB-453 and MDA-MB-231, luminal B type of breast cancer cell line T-47D, luminal A type of breast cancer cell line MCF-7, and human mammary epithelial cell line MCF-10A cells were purchased from Sino Biological (Beijing, China). RPMI 1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Biological Industries, Kibbutz Beit Haemek, Israel) was used to culture the cells at 37°C with 5% CO2.
PLASMID CONSTRUCTION AND TRANSFECTION:
The short hairpin RNA (shRNA) vector for THOR, overexpression vector for β-catenin and the responding empty vectors were constructed by GENEVIZ (Suzhou, Jiangsu, China), and denoted as THOR-shRNA (5′-CACCGGTCATGACCTGTGCATATG CCGAAGCATATGCACAGGTCATGACC-3′), negative control shRNA (5′-CACCCGACGTGCACCACGTGCTACTCGTACTCTTGATGCC GAGCACGGAA-3′), THOR-oe and β-catenin-oe, respectively. Lipofectamine 3000 (Thermo Fisher Scientific) was used for plasmid transfection according to the instructions. The corresponding empty vector acted as a control.
REAL-TIME QUANTITATIVE POLYMERASE CHAIN REACTION (RT-QPCR):
Total RNA was extracted by TRIzol (Tiangen, Beijing, China) and reverse transcribed using Takara reverse transcription kit (Tokyo, Japan). The target genes and control internal reference β-actin were amplified, respectively. The reaction conditions were 95°C for 15 minutes, 94°C for 15 seconds, 60°C for 30 seconds, 72°C for 60 seconds, 40 cycles, 95°C for 15 seconds, 60°C for 15 seconds, and 95°C for 15 seconds to verify the characteristics of the PCR products. Sequence Detector System 2.0 software was used to analyze the data and calculate the genes’ relative expressions.
WESTERN BLOT:
ProteinExt® Mammalian Total Protein Extraction Kit (Transgen, Beijing, China) was used to extract the total protein and protein concentration was quantitatively analyzed by BCA kit (Tiangen) as follows: take 1 million cells, discard the culture solution, wash twice with 1 mL precooled phosphate-buffered saline (PBS), centrifugate for 5 minutes at 3000 rpm, and carefully discard the supernatant. The supernatant was collected after centrifugation for 15 minutes. After sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS/PAGE), proteins were transferred to the polyvinylidene difluoride (PVDF) membrane (Merck Millipore, Billerica, MA, USA). 5% skimmed-milk powder was used to block the PVDF membrane for 1 hour, then we added the primary antibody: anti-Oct 1: 1000 (Cat# 11263-1-AP, Proteintech, Wuhan, China); anti-nanog 1: 1000 (Cat# 14295-1-AP, Proteintech); anti-CD44 1: 1000 (Cat# 15675-1-AP, Proteintech); anti-β-actin 1: 2000 (Cat# 60008-1-Ig, Proteintech); anti-β-catenin 1: 1000 (Cat# 17565-1-AP, Proteintech); anti-Sox9 1: 1000 (Cat# 67439-1-Ig, Proteintech); anti-c-Myc 1: 1000 (Cat# 10828-1-AP, Proteintech); anti-Axin2 1: 1000 (Cat# 20540-1-AP, Proteintech); anti-cyclin D1 1: 1000 (Cat# 26939-1-AP, Proteintech). Then we incubated at 4°C overnight. After washing, the secondary antibodies were added: HRP-conjugated Affinipure goat anti-mouse IgG(H+L) 1: 5000 (Cat# SA00001-1, Proteintech); HRP-conjugated Affinipure goat anti-rabbit IgG(H+L) 1: 5000 (Cat# SA00001-2, Proteintech) and then incubated for 1 hour at room temperature. ECL Plus reagent (Cat# PE0010, Solarbio, Beijing) was used to expose the membranes. The strips were quantitatively analyzed by GD5800 analysis system.
RNA IMMUNOPRECIPITATION (RIP):
The interaction between THOR and β-catenin protein was determined using a Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Merck Millipore) with anti-β-catenin and control IgG antibody.
:
The detailed procedure was referred to the previous study [19]. Briefly, BrU-labeled RNAs (THOR and THOR-anti-sense) were incubated with beads for another 2 hours at 4°C. Then, β-catenin RNA fragment was added into individual tubes and incubated at 4°C overnight. Subsequently, RNA was extracted from the supernatant after beads were digested using the miRNeasy kit (Qiagen, Duesseldorf), and β-catenin mRNA level was detected by qRT-PCR assay.
MRNA STABILITY ANALYSIS:
TNBC cells with THOR silencing or not was added with 5 μg/mL actinomycin D (ActD) to terminate transcription. Total RNA was extracted at 0, 2, 4, and 6 hours after adding ActD for quantitative analysis. The β-catenin mRNA half-life (tl/2) was measured compared to the mRNA level before adding ActD. The control group was the mRNA from cells treated with responding empty vector.
ALDH1 ACTIVITY ASSAY:
ALDH1 activity was evaluated using ALDEFLUOR™ Kit (Cat# KA3742, Stemcell Technologies) according to the instructions.
SPHERE-FORMATION ANALYSIS:
The cells in logarithmic phase were digested, centrifuged, the medium containing serum was removed, and washed twice with PBS. Then cells were suspended in serum-free DMEM/F12 medium containing 20 ng/mL bFGF (Sigma-Aldrich, St. Louis, MO, USA), 20 ng/mL EGF (Sigma), 20 μL/mL B27 (Sigma) and counted. The cells were cultured in the low adherent 6-well plates with 103 cells/well, and the spheres were calculated after 12 days. In detail, when the diameter of cell sphere was more than 100 μm, spheres size and number were measured and calculated by the confocal laser scanning microscope manually.
CELL VIABILITY ASSAY:
MDA-MB-231 and MDA-MB-435 cells in logarithmic growth period were digested by 0.25% trypsin to make single cell suspension, and the cell concentration was adjusted to 5×103/mL, and repeatedly to make the cells uniform and inoculate them in 96-well culture plates. Cells was added for culture with 5% CO2 at 37°C. The concentration of 5 mg/mL MTT solution (Yifeixue, Nanjing, China) was added on day 1, 2, and 3, respectively. The supernatant was removed after incubating at 37°C for 4 hours, and 150 μL dimethyl sulfoxide (DMSO) was added and shaken well to dissolve the crystal for 10 minutes. Then 570 nm was selected and the absorbance (OD) of each well was examined. The relative cell viability was calculated using the formula: cell survival rate=absorbance value of test group/absorbance value of control group×100%.
STATISTICAL ANALYSIS:
SPSS 22.0 software was used for statistical analysis. Results were denoted as the mean±standard deviation), single factor analysis of variance (ANOVA) was used for analysis, and non-paired
Results
LNCRNA THOR WAS OVEREXPRESSED IN TNBC:
THOR expression was initially examined in different sub-types of breast cancer tissues and it was identified that THOR was overexpressed in breast cancer tissues. Especially, THOR displayed the highest level in TNBC tissues (Figure 1A). Additionally, THOR expression was evaluated in normal breast epithelial and different types of breast cancer cells. As shown in Figure 1B, THOR displayed the highest level in TNBC cells and was significantly overexpressed in breast cancer cells. Since the proportion of CSCs is significantly higher in TNBC than that in other sub-types, we assumed that THOR could regulate TNBC cell stemness.
THOR PROMOTED THE STEMNESS OF TNBC CELLS:
We then knocked down THOR expression in TNBC cells to explore its roles as THOR holds the highest level in TNBC cells. The knockdown efficiency was confirmed by RT-qPCR assay (Figure 2A). Notably, it was found that THOR knockdown had no effects on TNBC cell viability (Figure 2B, 2C). Then we determined its effects on TNBC cell stemness. As shown in Figure 2D–2F, the expression of CSC regulatory factors (Oct4, CD44, and Nanog) was decreased by THOR knockdown in TNBC cells. Additionally, ALDH1 activity was reduced by THOR knockdown too (Figure 2G). Moreover, the sphere-formation ability was suppressed by THOR knockdown, as evidenced by decreased sphere size and number (Figures 2G–2I). On the contrary, THOR overexpression significantly increased the stemness of TNBC cells, which was evident as the increased sphere-formation ability and stemness gene expression (Figure 3). These results suggest that THOR could enhance TNBC cell stemness.
THOR DIRECTLY BOUND TO β-CATENIN MRNA, ENHANCES ITS MRNA STABILITY AND EXPRESSION:
Then we explored the underlying mechanisms contributing to THOR-mediated effects on TNBC cell stemness. Previous studies have shown that THOR promotes CSC expansion in liver cancer through regulating β-catenin signaling [21], and osteosarcoma by stablizing SOX9 mRNA [20]. Thus, we wonder which signaling pathway was involved, or whether both pathways were involved in THOR-induced effects on TNBC cell stemness. The expression of SOX9 and β-catenin was examined in TNBC cells with THOR silencing or not and it was shown that the expression of β-catenin but not SOX9 was decreased by THOR knockdown (Figure 4A–4C). Therefore, we speculate that THOR may promote TNBC cell stemness at least by activating β-catenin signaling. As lncRNA could directly bind to RNAs or proteins, we initially examined whether THOR physically interacts with β-catenin mRNA using the RNA-RNA in vitro interaction assay, and β-catenin protein using RIP assay, it was found that THOR directly bound to β-catenin mRNA but not β-catenin protein (Figure 4D, 4E). Additionally, the mRNA stability assay on β-catenin mRNA showed that THOR knockdown reduced β-catenin mRNA stability, evident by the decrease of β-catenin mRNA half-life (Figure 4F, 4G). Furthermore, the activity of TOPFlash luciferase reporter (a responsive vector of β-catenin) was reduced by THOR knockdown, while the nonresponsive FOPFlash control was unaffected (Figure 4H). In addition, the protein expression of β-catenin and its downstream effectors (c-Myc, AXIN2, and cyclin-D1) was decreased by THOR knockdown in TNBC cells (Figure 4I). Thus, our results demonstrated that THOR could activate β-catenin signaling through directly binding to β-catenin mRNA but not β-catenin protein.
THOR REGULATED TNBC CELL STEMNESS DEPENDENT ON β-CATENIN EXPRESSION:
Finally, we explored whether THOR regulates TNBC cell stemness through β-catenin. β-catenin was overexpressed in TNBC cells with or without THOR knockdown (Figure 5A, 5B). As expected, THOR knockdown-mediated decrease of stemness marker expression was partially reversed by β-catenin overexpression (Figures 5C–5F). Additionally, β-catenin overexpression rescued the reduced ALDH1 activity led by THOR knockdown (Figure 5G). Furthermore, the decreased sphere-formation capacity resulted by THOR silencing was diminished by β-catenin overexpression (Figure 5H, 5I). Collectively, these results indicate that THOR regulated TNBC cell stemness dependent on β-catenin expression.
Discussion
Most of the human genome consists of non-coding sequences, with less than 2% protein-coding genes, which suggests the importance of non-coding RNA in human life [4]. Because of its low expression and poor evolutionary conservativeness, lncRNA was once considered a “transcriptional noise” in chromosomes and neglected [22]. In recent years, with the rapid development of high-throughput sequencing and chip technology, the importance of lncRNA in organisms has gradually been taken seriously. More and more studies have shown that lncRNA plays a role similar to proto-oncogene or anti-oncogene in many malignant tumors. In this study, we focused on THOR roles in TNBC cell stemness since its roles in breast cancer has never been found and it is found at its highest level in TNBC. We found that THOR could not affect TNBC cell proliferation but could significantly attenuate TNBC cell stemness, as evidenced by decreased sphere-formation ability, ALDH1 activity, and CSC regulatory factor expression. To the best of our knowledge, this is the first study showing THOR effects in breast cancer progression.
β-catenin signaling is one of the important pathways in tumor stemness [23]. Previous studies have indicated that THOR exerts its effects on tumor stemness through regulating Sox9 or β-catenin signaling in different tumors individually [19,20]. In the present study, both Sox9 and β-catenin expression were determined in TNBC cells with THOR knockdown and we found that β-catenin expression but not Sox9 expression was decreased by THOR knockdown, which indicates that THOR functions through β-catenin signaling but not Sox9 signaling, and acts through different mechanisms in tumors. Additionally, although the promoting effects of THOR on β-catenin signaling have been confirmed before, the detailed mechanisms by which THOR regulating β-catenin expression is still unclear. THOR has been confirmed to be located in cytoplasm and lncRNA localized in cytoplasm mainly affects gene expression in the following ways: 1) increase the stability of RNA, such as lncRNA-TINCR can stabilize STAU1-RNA through interacting with TINCR box motif of STAU1-RNA. 2) Promote the degradation of STAU1-RNA, such as the lncRNA encoded by 1/2-sbsRNAs can promote the degradation of STAU1-mRNA by binding to the Alu reaction elements in its 3′UTR region [24]. 3) Participate in stress conditions, such as the translocation of antisense lncRNA of Uchl1 from nucleus to cytoplasm, and binds to the 5′end of Uchl1-mRNA to promote the transcription of Uchl1 [25]. 4) Inhibit protein translation by direct interaction with RNA, such as lincRNA-p21 can bind directly to RcK mRNA and inhibit its RcK entering ribosome for protein translation [26]. In the current work, we revealed that THOR could physically interact with β-catenin mRNA through performing
Conclusions
THOR may be a key lncRNA molecule regulating TNBC occurrence and development. Further study on THOR roles in TNBC may provide new ideas for TNBC diagnosis and treatment.
Figures
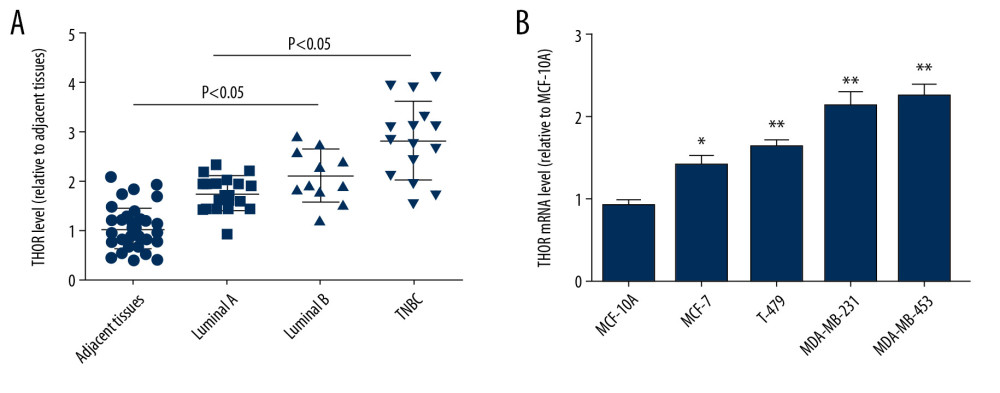 Figure 1. LncRNA THOR is overexpressed in TNBC tissues and cells. (A) RT-qPCR was used to examine THOR level in different types of breast cancer and adjacent tissues. (B) RT-qPCR was conducted to determine THOR level in different types of breast cancer and normal breast epithelial cells. * P<0.05, ** P<0.01. lncRNA – long non-coding RNA; TBBC – triple negative breast cancer; RT-q-PCR – real-time quantitative polymerase chain reaction.
Figure 1. LncRNA THOR is overexpressed in TNBC tissues and cells. (A) RT-qPCR was used to examine THOR level in different types of breast cancer and adjacent tissues. (B) RT-qPCR was conducted to determine THOR level in different types of breast cancer and normal breast epithelial cells. * P<0.05, ** P<0.01. lncRNA – long non-coding RNA; TBBC – triple negative breast cancer; RT-q-PCR – real-time quantitative polymerase chain reaction. 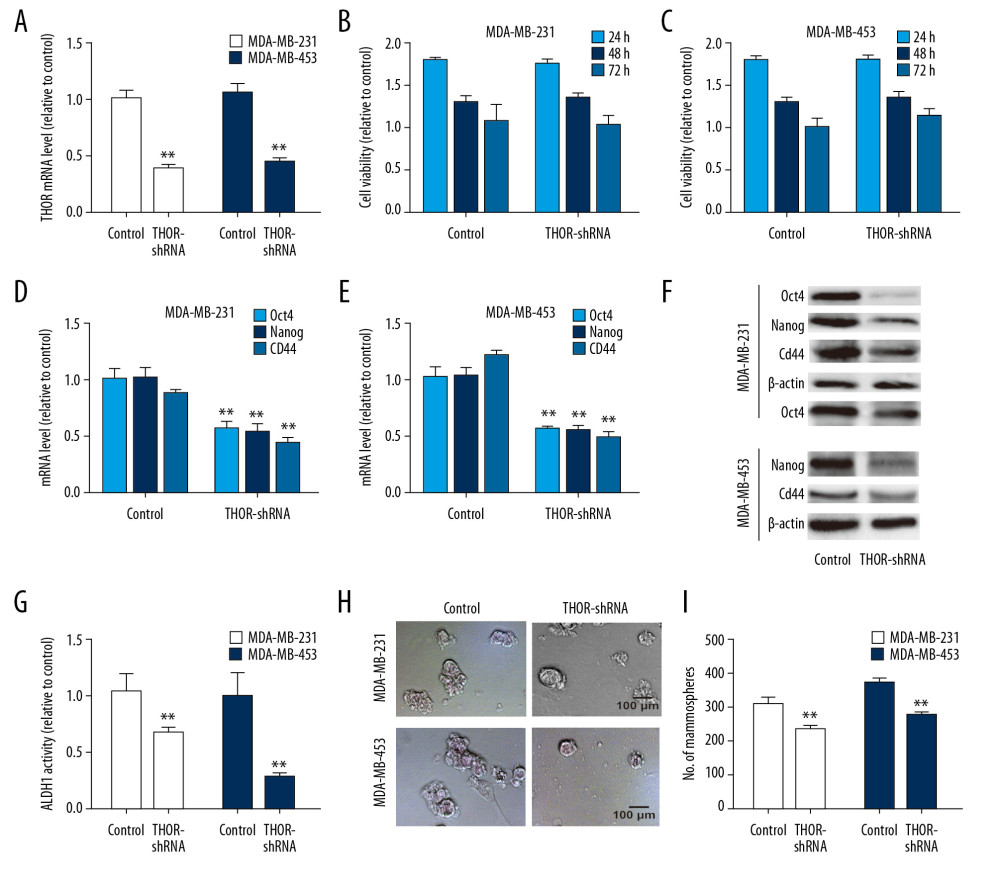 Figure 2. THOR knockdown reduces the stemness of TNBC cells. (A) RT-qPCR was constructed to detect the knockdown efficiency of THOR-shRNA in TNBC cells. (B, C) The effects of THOR-shRNA on TNBC cell viability. (D–F) The mRNA and protein levels of stemness markers were examined in TNBC cells with THOR knockdown or not. (G) ALDH1 activity was measured in TNBC cells depicted in (D). (H, I) The capacity of sphere-formation was evaluated in TNBC cells described in (D). ** P<0.01. TNBC – triple negative breast cancer; RT-q-PCR – real-time quantitative polymerase chain reaction; shRNA – short hairpin RNA; mRNA – messenger RNA.
Figure 2. THOR knockdown reduces the stemness of TNBC cells. (A) RT-qPCR was constructed to detect the knockdown efficiency of THOR-shRNA in TNBC cells. (B, C) The effects of THOR-shRNA on TNBC cell viability. (D–F) The mRNA and protein levels of stemness markers were examined in TNBC cells with THOR knockdown or not. (G) ALDH1 activity was measured in TNBC cells depicted in (D). (H, I) The capacity of sphere-formation was evaluated in TNBC cells described in (D). ** P<0.01. TNBC – triple negative breast cancer; RT-q-PCR – real-time quantitative polymerase chain reaction; shRNA – short hairpin RNA; mRNA – messenger RNA. 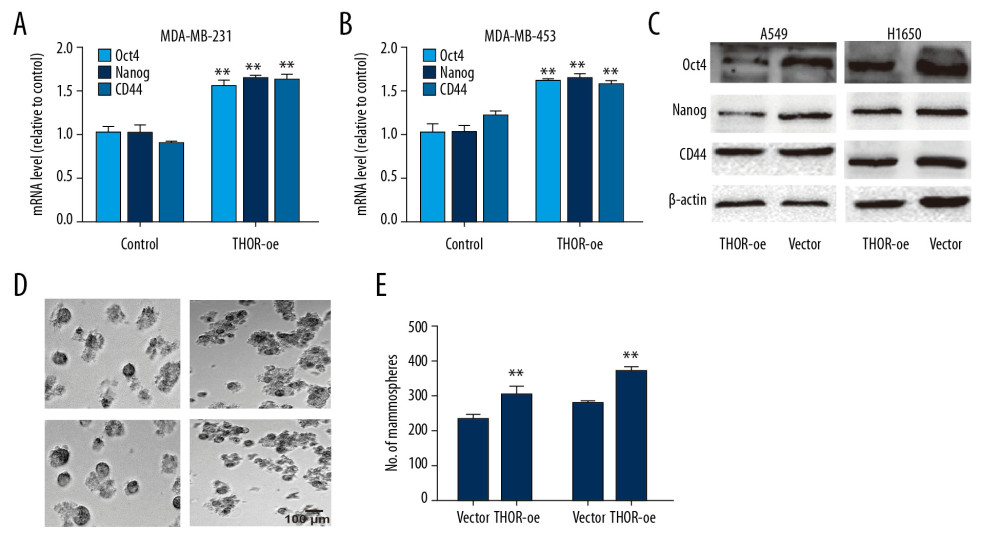 Figure 3. THOR overexpression promotes the stemness of TNBC cells. (A–C) The mRNA and protein expression of stemness genes were determined in TNBC cells with THOR overexpression or not. (D, E) The sphere size and number were examined in TNBC cells with THOR overexpression or not.** P<0.01. TNBC,– triple negative breast cancer; mRNA – messenger RNA.
Figure 3. THOR overexpression promotes the stemness of TNBC cells. (A–C) The mRNA and protein expression of stemness genes were determined in TNBC cells with THOR overexpression or not. (D, E) The sphere size and number were examined in TNBC cells with THOR overexpression or not.** P<0.01. TNBC,– triple negative breast cancer; mRNA – messenger RNA. 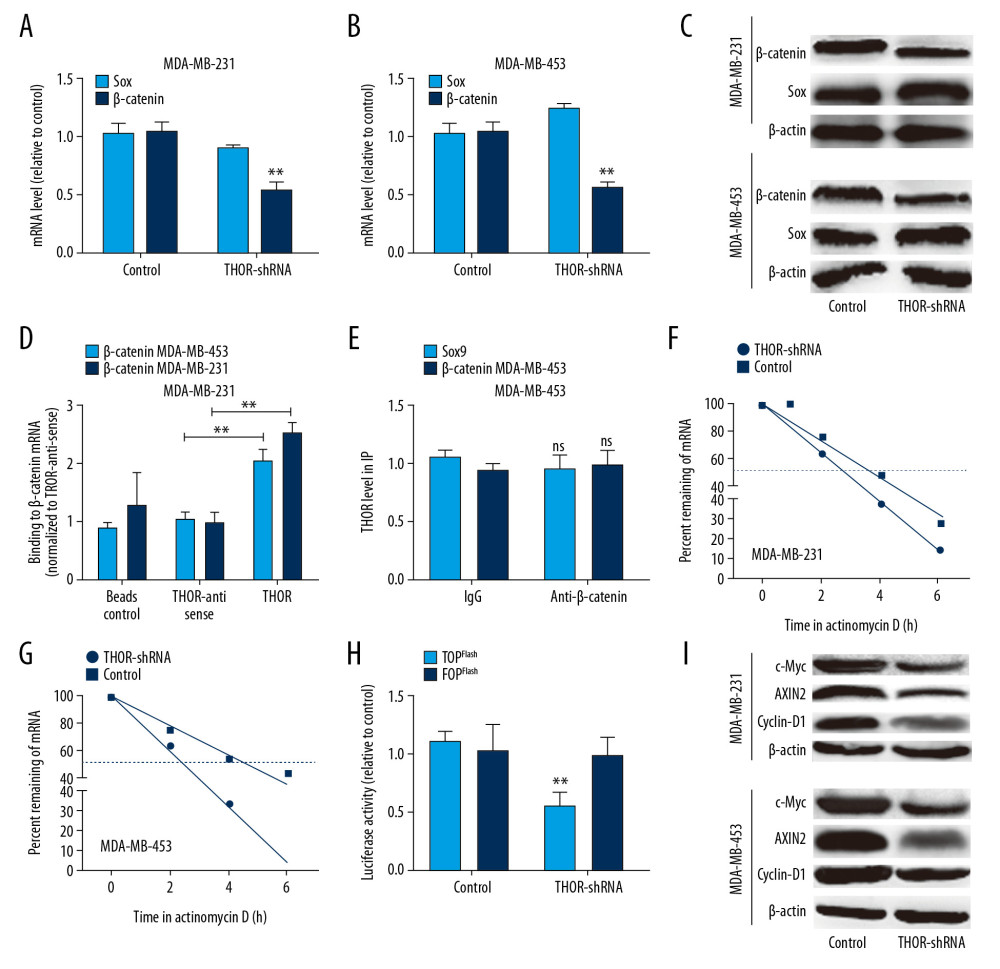 Figure 4. THOR directly binds to β-catenin mRNA, enhances its mRNA stability and expression. (A–C) The mRNA and protein levels of Sox9 and β-catenin were examined in TNBC cells with THOR knockdown or not. (D) The in vitro RNA-RNA interaction was performed to examine the THOR-β-catenin RNA interaction in TNBC cells. (E) RIP assay on THOR level in RNA pulled down by anti-β-catenin. (F, G) Analysis on the mRNA stability of β-catenin was performed in TNBC cells with THOR knockdown or not. (H) Luciferase reporter analysis on the TOPflash and FOPflash in MDA-MB-453 cells with THOR knockdown or not. (I) The expression of the downstream effectors of β-catenin was detected in TNBC cells with THOR knockdown or not. ** P<0.01. mRNA – messenger RNA; TNBC – triple negative breast cancer; RIP – RNA immunoprecipitation.
Figure 4. THOR directly binds to β-catenin mRNA, enhances its mRNA stability and expression. (A–C) The mRNA and protein levels of Sox9 and β-catenin were examined in TNBC cells with THOR knockdown or not. (D) The in vitro RNA-RNA interaction was performed to examine the THOR-β-catenin RNA interaction in TNBC cells. (E) RIP assay on THOR level in RNA pulled down by anti-β-catenin. (F, G) Analysis on the mRNA stability of β-catenin was performed in TNBC cells with THOR knockdown or not. (H) Luciferase reporter analysis on the TOPflash and FOPflash in MDA-MB-453 cells with THOR knockdown or not. (I) The expression of the downstream effectors of β-catenin was detected in TNBC cells with THOR knockdown or not. ** P<0.01. mRNA – messenger RNA; TNBC – triple negative breast cancer; RIP – RNA immunoprecipitation. 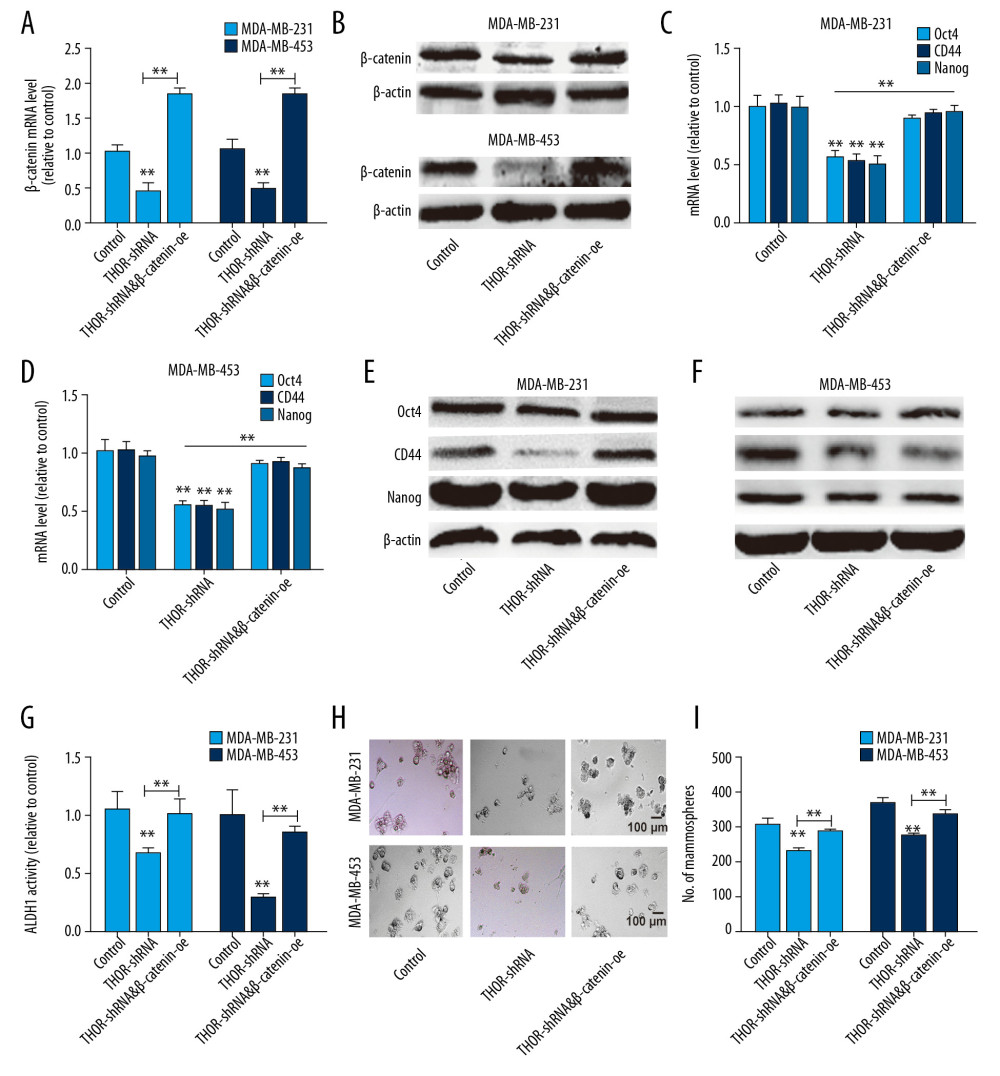 Figure 5. THOR regulates TNBC cell stemness dependent on β-catenin expression. (A, B) β-catenin mRNA and protein levels were determined in TNBC cells with THOR knockdown or not, as well as β-catenin overexpression. (C–F) The analysis on the expression of stemness markers in the cells denoted in (B). (G) ALDH1 activity was assessed in the cells denoted in (B). (H, I) The determination on sphere-formation ability in the cells denoted in (B). ** P<0.01. TNBC – triple negative breast cancer; mRNA – messenger RNA.
Figure 5. THOR regulates TNBC cell stemness dependent on β-catenin expression. (A, B) β-catenin mRNA and protein levels were determined in TNBC cells with THOR knockdown or not, as well as β-catenin overexpression. (C–F) The analysis on the expression of stemness markers in the cells denoted in (B). (G) ALDH1 activity was assessed in the cells denoted in (B). (H, I) The determination on sphere-formation ability in the cells denoted in (B). ** P<0.01. TNBC – triple negative breast cancer; mRNA – messenger RNA. References
1. Lima ZS, Ghadamzadeh M, Arashloo FT, Recent advances of therapeutic targets based on the molecular signature in breast cancer: Genetic mutations and implications for current treatment paradigms: J Hematol Oncol, 2019; 12(1); 38
2. Han Y, Xie W, Song DG, Powell DJ: J Hematol Oncol, 2018; 11(1); 92
3. Zheng L, Xiang C, Li X, STARD13-correlated ceRNA network-directed inhibition on YAP/TAZ activity suppresses stemness of breast cancer via co-regulating Hippo and Rho-GTPase/F-actin signaling: J Hematol Oncol, 2018; 11(1); 72
4. Wang J, Xie S, Yang J, The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy: J Hematol Oncol, 2019; 12(1); 81
5. Zheng L, Meng X, Li X, miR-125a-3p inhibits ERalpha transactivation and overrides tamoxifen resistance by targeting CDK3 in estrogen receptor-positive breast cancer: FASEB J, 2018; 32(2); 588-600
6. Lin Y, Guo W, Li N, Polymorphisms of long non-coding RNA HOTAIR with breast cancer susceptibility and clinical outcomes for a southeast Chinese Han population: Oncotarget, 2018; 9(3); 3677-89
7. Chou J, Wang B, Zheng T, MALAT1 induced migration and invasion of human breast cancer cells by competitively binding miR-1 with cdc42: Biochem Biophys Res Commun, 2016; 472(1); 262-69
8. Mansoori Y, Tabei MB, Askari A, Expression levels of breast cancer-related GAS5 and LSINCT5 lncRNAs in cancer-free breast tissue: Molecular associations with age at menarche and obesity: Breast J, 2018; 24(6); 876-82
9. Singh R, Gupta SC, Peng WX, Regulation of alternative splicing of Bcl-x by BC200 contributes to breast cancer pathogenesis: Cell Death Dis, 2016; 7(6); e2262
10. Xing F, Liu Y, Wu SY, Loss of XIST in breast cancer activates MSN-c-Met and reprograms microglia via exosomal miRNA to promote brain metastasis: Cancer Res, 2018; 78(15); 4316-30
11. Li W, Zhai L, Wang H, Downregulation of lncRNA GAS5 causes trastuzumab resistance in breast cancer: Oncotarget, 2016; 7(19); 27778-86
12. Han L, Zhang HC, Li L, Downregulation of long noncoding RNA HOTAIR and EZH2 induces apoptosis and inhibits proliferation, invasion, and migration of human breast cancer cells: Cancer Biother Radiopharm, 2018; 33(6); 241-51
13. Berteaux N, Lottin S, Monte D, H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1: J Biol Chem, 2005; 280(33); 29625-36
14. Yang T, Li S, Liu J, LncRNA-NKILA/NF-kappaB feedback loop modulates laryngeal cancer cell proliferation, invasion, and radioresistance: Cancer Med, 2018; 7(5); 2048-63
15. Hosono Y, Niknafs YS, Prensner JR, Oncogenic role of THOR, a conserved cancer/testis long non-coding RNA: Cell, 2017; 171(7); 1559-72.e20
16. Chen W, Chen M, Xu Y: Biochem Biophys Res Commun, 2018; 499(4); 913-19
17. Ye XT, Huang H, Huang WP, Hu WL, LncRNA THOR promotes human renal cell carcinoma cell growth: Biochem Biophys Res Commun, 2018; 501(3); 661-67
18. Cheng Z, Lei Z, Yang P, Long non-coding RNA THOR promotes cell proliferation and metastasis in hepatocellular carcinoma: Gene, 2018; 678; 129-36
19. Song H, Xu Y, Shi L, LncRNA THOR increases the stemness of gastric cancer cells via enhancing SOX9 mRNA stability: Biomed Pharmacother, 2018; 108; 338-46
20. Wu H, He Y, Chen H, LncRNA THOR increases osteosarcoma cell stemness and migration by enhancing SOX9 mRNA stability: FEBS Open Bio, 2019; 9(4); 781-90
21. Cheng Z, Lei Z, Yang P, Long non-coding RNA THOR promotes liver cancer stem cells expansion via beta-catenin pathway: Gene, 2019; 684; 95-103
22. Zheng L, Li X, Gu Y, The 3′UTR of the pseudogene CYP4Z2P promotes tumor angiogenesis in breast cancer by acting as a ceRNA for CYP4Z1: Breast Cancer Res Treat, 2015; 150(1); 105-18
23. El Kharbili M, Agaesse G, Barbollat-Boutrand L, Tspan8-beta-catenin positive feedback loop promotes melanoma invasion: Oncogene, 2019; 38(20); 3781-93
24. Gong C, Maquat LE, LncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements: Nature, 2011; 470(7333); 284-88
25. Liu H, Povysheva N, Rose ME, Role of UCHL1 in axonal injury and functional recovery after cerebral ischemia: Proc Natl Acad Sci USA, 2019; 116(1); 4643-50
26. Chen Y, Wei G, Xia H, Down regulation of lincRNA-p21 contributes to gastric cancer development through Hippo-independent activation of YAP: Oncotarget, 2017; 8(38); 63813-24
27. Xue J, Zhong S, Sun BM, Lnc-THOR silencing inhibits human glioma cell survival by activating MAGEA6-AMPK signaling: Cell Death Dis, 2019; 10(11); 866
28. Yang H, Fu G, Liu F, LncRNA THOR promotes tongue squamous cell carcinomas by stabilizing IGF2BP1 downstream targets: Biochimie, 2019; 165; 9-18
Figures
 Figure 1. LncRNA THOR is overexpressed in TNBC tissues and cells. (A) RT-qPCR was used to examine THOR level in different types of breast cancer and adjacent tissues. (B) RT-qPCR was conducted to determine THOR level in different types of breast cancer and normal breast epithelial cells. * P<0.05, ** P<0.01. lncRNA – long non-coding RNA; TBBC – triple negative breast cancer; RT-q-PCR – real-time quantitative polymerase chain reaction.
Figure 1. LncRNA THOR is overexpressed in TNBC tissues and cells. (A) RT-qPCR was used to examine THOR level in different types of breast cancer and adjacent tissues. (B) RT-qPCR was conducted to determine THOR level in different types of breast cancer and normal breast epithelial cells. * P<0.05, ** P<0.01. lncRNA – long non-coding RNA; TBBC – triple negative breast cancer; RT-q-PCR – real-time quantitative polymerase chain reaction. Figure 2. THOR knockdown reduces the stemness of TNBC cells. (A) RT-qPCR was constructed to detect the knockdown efficiency of THOR-shRNA in TNBC cells. (B, C) The effects of THOR-shRNA on TNBC cell viability. (D–F) The mRNA and protein levels of stemness markers were examined in TNBC cells with THOR knockdown or not. (G) ALDH1 activity was measured in TNBC cells depicted in (D). (H, I) The capacity of sphere-formation was evaluated in TNBC cells described in (D). ** P<0.01. TNBC – triple negative breast cancer; RT-q-PCR – real-time quantitative polymerase chain reaction; shRNA – short hairpin RNA; mRNA – messenger RNA.
Figure 2. THOR knockdown reduces the stemness of TNBC cells. (A) RT-qPCR was constructed to detect the knockdown efficiency of THOR-shRNA in TNBC cells. (B, C) The effects of THOR-shRNA on TNBC cell viability. (D–F) The mRNA and protein levels of stemness markers were examined in TNBC cells with THOR knockdown or not. (G) ALDH1 activity was measured in TNBC cells depicted in (D). (H, I) The capacity of sphere-formation was evaluated in TNBC cells described in (D). ** P<0.01. TNBC – triple negative breast cancer; RT-q-PCR – real-time quantitative polymerase chain reaction; shRNA – short hairpin RNA; mRNA – messenger RNA. Figure 3. THOR overexpression promotes the stemness of TNBC cells. (A–C) The mRNA and protein expression of stemness genes were determined in TNBC cells with THOR overexpression or not. (D, E) The sphere size and number were examined in TNBC cells with THOR overexpression or not.** P<0.01. TNBC,– triple negative breast cancer; mRNA – messenger RNA.
Figure 3. THOR overexpression promotes the stemness of TNBC cells. (A–C) The mRNA and protein expression of stemness genes were determined in TNBC cells with THOR overexpression or not. (D, E) The sphere size and number were examined in TNBC cells with THOR overexpression or not.** P<0.01. TNBC,– triple negative breast cancer; mRNA – messenger RNA. Figure 4. THOR directly binds to β-catenin mRNA, enhances its mRNA stability and expression. (A–C) The mRNA and protein levels of Sox9 and β-catenin were examined in TNBC cells with THOR knockdown or not. (D) The in vitro RNA-RNA interaction was performed to examine the THOR-β-catenin RNA interaction in TNBC cells. (E) RIP assay on THOR level in RNA pulled down by anti-β-catenin. (F, G) Analysis on the mRNA stability of β-catenin was performed in TNBC cells with THOR knockdown or not. (H) Luciferase reporter analysis on the TOPflash and FOPflash in MDA-MB-453 cells with THOR knockdown or not. (I) The expression of the downstream effectors of β-catenin was detected in TNBC cells with THOR knockdown or not. ** P<0.01. mRNA – messenger RNA; TNBC – triple negative breast cancer; RIP – RNA immunoprecipitation.
Figure 4. THOR directly binds to β-catenin mRNA, enhances its mRNA stability and expression. (A–C) The mRNA and protein levels of Sox9 and β-catenin were examined in TNBC cells with THOR knockdown or not. (D) The in vitro RNA-RNA interaction was performed to examine the THOR-β-catenin RNA interaction in TNBC cells. (E) RIP assay on THOR level in RNA pulled down by anti-β-catenin. (F, G) Analysis on the mRNA stability of β-catenin was performed in TNBC cells with THOR knockdown or not. (H) Luciferase reporter analysis on the TOPflash and FOPflash in MDA-MB-453 cells with THOR knockdown or not. (I) The expression of the downstream effectors of β-catenin was detected in TNBC cells with THOR knockdown or not. ** P<0.01. mRNA – messenger RNA; TNBC – triple negative breast cancer; RIP – RNA immunoprecipitation. Figure 5. THOR regulates TNBC cell stemness dependent on β-catenin expression. (A, B) β-catenin mRNA and protein levels were determined in TNBC cells with THOR knockdown or not, as well as β-catenin overexpression. (C–F) The analysis on the expression of stemness markers in the cells denoted in (B). (G) ALDH1 activity was assessed in the cells denoted in (B). (H, I) The determination on sphere-formation ability in the cells denoted in (B). ** P<0.01. TNBC – triple negative breast cancer; mRNA – messenger RNA.
Figure 5. THOR regulates TNBC cell stemness dependent on β-catenin expression. (A, B) β-catenin mRNA and protein levels were determined in TNBC cells with THOR knockdown or not, as well as β-catenin overexpression. (C–F) The analysis on the expression of stemness markers in the cells denoted in (B). (G) ALDH1 activity was assessed in the cells denoted in (B). (H, I) The determination on sphere-formation ability in the cells denoted in (B). ** P<0.01. TNBC – triple negative breast cancer; mRNA – messenger RNA. In Press
06 Mar 2024 : Clinical Research
Comparison of Outcomes between Single-Level and Double-Level Corpectomy in Thoracolumbar Reconstruction: A ...Med Sci Monit In Press; DOI: 10.12659/MSM.943797
21 Mar 2024 : Meta-Analysis
Economic Evaluation of COVID-19 Screening Tests and Surveillance Strategies in Low-Income, Middle-Income, a...Med Sci Monit In Press; DOI: 10.12659/MSM.943863
10 Apr 2024 : Clinical Research
Predicting Acute Cardiovascular Complications in COVID-19: Insights from a Specialized Cardiac Referral Dep...Med Sci Monit In Press; DOI: 10.12659/MSM.942612
06 Mar 2024 : Clinical Research
Enhanced Surgical Outcomes of Popliteal Cyst Excision: A Retrospective Study Comparing Arthroscopic Debride...Med Sci Monit In Press; DOI: 10.12659/MSM.941102
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952








