07 May 2020: Lab/In Vitro Research
Potential Molecular Mechanisms and Drugs for Aconitine-Induced Cardiotoxicity in Zebrafish through RNA Sequencing and Bioinformatics Analysis
Mingzhu Wang1CDEF, Yanan Shi1BDF, Lei Yao1CDF, Qiang Li2ACDFG, Youhua Wang3ABDFG, Deyu Fu1ABEFG*DOI: 10.12659/MSM.924092
Med Sci Monit 2020; 26:e924092
Abstract
BACKGROUND: Accumulating evidence suggests that cardiotoxicity is one of the main manifestations of aconitine (AC) poisoning. However, the molecular mechanism of AC-induced cardiotoxicity remains unclear, there is little direct evidence for therapeutic targets and drugs of AC-induced cardiotoxicity.
MATERIAL AND METHODS: Zebrafish were exposed to AC to evaluate cardiotoxicity by calculating the heart rates and observing the changes of cardiac and vascular structure. RNA-seq (RNA sequencing) and bioinformatics analysis were used to obtain differentially expressed genes (DEGs). The anti-AC cardiotoxicity compound was identified via connectivity map (CMAP) analysis and molecular docking.
RESULTS: AC-induced cardiotoxicity in zebrafish predominantly included arrhythmias, extended sinus venous and bulbus arteriosus (SV-BA) distance, and larger pericardial edema aera. A total of 1380 DEGs were identified by RNA-seq and bioinformatics analysis. cyclin-dependent kinase-1 (CDK1) was screened as the hub gene and the most potential therapeutic target due to its significant downregulation in cardiotoxicity based on protein-protein interaction (PPI) and drug-gene interaction (DGIdb) network analysis. Cell cycle signal pathway was the most significant pathways identified in the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis. Furthermore, the expression of CDK1 was validated in the Gene Expression Omnibus (GEO) database GSE71906, GSE65705, and GSE95140. Finally, heptaminol was identified as a novel anti-AC cardiotoxicity compound via CMAP analysis and molecular docking.
CONCLUSIONS: Totally, hub genes and key pathways identified in this study can aid in the understanding of the molecular changes in AC-induced cardiotoxicity. Meanwhile, we provide a systematic method to explore drug toxicity prevention and treatment.
Keywords: Aconitine, Drug-Related Side Effects and Adverse Reactions, Sequence Analysis, RNA, Zebrafish, Biomarkers, Tumor, cardiotoxicity, Computational Biology, Databases, Genetic, Gene Expression Profiling, gene ontology, Gene Regulatory Networks, Heart Rate, Molecular Docking Simulation, Protein Interaction Mapping, Protein Interaction Maps, Whole Exome Sequencing
Background
Aconitum carmichaelii Debeaux (Fuzi in Chinese) has been frequently employed in traditional Chinese medicine for thousands of years to treat inflammation, pain, neurologic diseases and cardiovascular diseases [1,2]. With the development of pharmacological research, aconitine (AC) has been identified as one of the main active compounds in aconitum with a variety of pharmacological activities such as analgesia and anti-inflammatory and anti-tumor activities [3,4]. However, the clinical efficacy and toxicity features of AC coexist [5], which limits its clinical application [6]. In addition, the treatment outcome for AC poisoning is unsatisfactory because of limitations in the antidote for AC poisoning [7].
Cardiotoxicity is one of the most prominent lethal causes of acute AC poisoning [8]. The typical symptoms of cardiovascular manifestations of acute AC poisoning include hypotensive and bradycardic actions, palpitations, chest pains, sinus or ventricular tachycardia, and ventricular fibrillation [9–11]. Current research on AC cardiotoxicity has mainly focused on arrhythmia and blood pressure, and the discussion of the mechanism of cardiotoxic effects has mainly focused on the regulation of ion channels and energy metabolism, and oxidative stress injury [12–14]. However, the toxicity of AC to the cardiac structure has rarely been explored and the specific mechanism is not well understood, and thus needs further study. Based on certain advantages of zebrafish for cardiovascular studies and the development of high-throughput sequencing technologies, we investigated the cardiac structural toxicity of AC and the underlying molecular mechanism.
The zebrafish is one of the most important vertebrate models used to study the mechanism of toxicology for cardiovascular disease [15]. Firstly, the hearts of zebrafish can be imaged under the microscope because their embryos and the young fish bodies are transparent, so the heart rates and blood circulation are visible and easy to observe. Secondly, the zebrafish share similar physiological and biochemical characteristics with mammals. The similar genome between humans and zebrafish is up to 87%, therefore it can be applied to investigate the molecular mechanism of human diseases or high-throughput drug screening [16]. Thirdly, cardiac fluorescence transgenic (cmlc2: GFP) zebrafish emit green under a fluorescence microscope, which makes it accurate and convenient to observe its heart structure changes [17]. Taken together, zebrafish have certain benefits for studying the effect of drugs on cardiovascular disease.
With the maturity and development of transcriptome sequencing technology, high-throughput platforms have been widely applied for gene expression analysis, especially in the study on pharmacology and toxicology of different herb monomers [18,19]. However, the data from high-throughput sequencing is huge, so it requires analysis by bioinformatics technology [20]. Screening for hub genes and key signal pathways played a key role in pharmacology and toxicology studies.
In the field of biomedical research, the ultimate objective is to link human diseases to the genes that clarify them and drugs that cure them. A druggable genome can be used to predict the drug-gene interactions, which are helpful to identify drugs that might inhibit or otherwise modulate those genes. The DGIdb (drug-gene interaction) and CMAP (connectivity map) databases provide the potential to discover the relationship among drugs, genes, and diseases [21,22], and there have been many studies on therapeutic targets and drugs for diseases screened from these databases, including ischemic stroke, obesity, osteonecrosis, and cancer [23–25].
The purpose of this study was to verify the cardiotoxicity of AC and investigate the potential therapeutic targets, mechanisms, and drugs. In this work, the zebrafish was used as a vertebrate model for evaluating the AC-induced cardiotoxicity by observing the changes in heart rates, cardiac morphology, and the sinus venous and bulbus arteriosus (SV-BA) distance. Furthermore, RNA-seq was applied to detect the differentially expressed genes (DEGs) of AC-induced cardiotoxicity in zebrafish. Next, hub therapeutic targets and key signaling pathways were obtained via bioinformatics and network analysis of DEGs. Finally, we identify the prospective drug for AC-induced cardiotoxicity through the DGIdb and CMAP databases and verify the relationship between drugs and targets by molecular docking.
Material and Methods
CHEMICALS AND REAGENTS:
Aconitine with purity >98% was bought from Chengdu Cdmust Bio-Technology Co., Ltd. (Chengdu, China). Dimethyl sulfoxide (DMSO) were obtained from WAK-Chemie (Steinbach, Germany) to facilitate dissolution.
ESTABLISHMENT OF THE ZEBRAFISH MODEL:
Zebrafish, transgenic (cmlc2: GFP) Tübingen (TU) strain, belonging to the Danio family of Cyprinidae, were provided by the Institute of Pediatrics, Children’s Hospital of Fudan University. The heart of a young zebrafish glows green under a fluorescence microscope, which facilitates the observation of zebrafish hearts and the measurement of related indicators. Adult zebrafish were bred in a water temperature of 28°C, at a pH of 7.0 to 7.5, and conductivity of 400–600 μs/cm. Zebrafish were fed 3 times a day, and exposed to fluorescent light for 14 hours and to darkness for 10 hours [26]. All procedures were approved by the Children’s Hospital of Fudan University.
CARDIOTOXICITY EVALUATION OF AC ON ZEBRAFISH:
The embryos were submerged in culture medium with cautious use of a needle and placed in an incubator. A previous study showed that at 72 hours post-fertilization (hpf), the half-maximal effective concentration (EC50) of the cardiac toxicity of AC to zebrafish embryos was 14.49 mg/L. AC has even been used as the method to prepare cardiac arrhythmic models in modern heart research [27,28]. At 48 hpf, the embryos were divided into an AC 15 mg/L group and a Control group (contained cultivated water with 0.2% DMSO). The timing of drug intervention ranged from 48 hpf to 72 hpf in zebrafish.
The embryos were subsequently adjusted to a side-lying position with cautious use of a needle. Ten embryos were collected in each treatment group. The heartbeats of the embryos were recorded by counting beats per minute on a live video, as previously described [29], and SV-BA distance, which could reflect the drug influence on the heart of zebrafish quantitatively [30], was measured by ImageJ (http://www.rsb.info.nih.gov/ij/) as previously described [31]. Images and videos were recorded at 48 hpf and 72 hpf.
RNA SEQUENCING:
To assess the potential effects of AC on the heart of zebrafish, embryos were exposed to system water or AC 15 mg/L (20 embryos per Petri dish) from 48 to 72 hpf as described. All samples were stored at −80°C until total RNA extraction. Total RNA extraction was performed with TRIZol reagents from Invitrogen following the manufacturer’s instructions. RNA library construction was then performed by BGI Co., Ltd., Shenzhen, China (http://www.genomics.cn/) and specific steps were followed using the methods previously described [32].
BIOINFORMATIC AND NETWORK ANALYSIS OF RNA-SEQ DATA:
Raw data from Illumina HiSeq sequencing were filtered, and clean reads were matched to the reference sequence. Quantitative analysis was conducted on the known genes, the PossionDis algorithm was applied to perform differential gene detection and screened |log2(FoldChange)| >1&qvalue <0.001 as the DEGs. Then the DEGs were analyzed by Database for Annotation, Visualization and Integrated Discovery (DAVID) database (https://david.ncifcrf.gov/) to explore the Gene Ontology (GO) functional annotation and the Kyoto Encyclopedia of Genes and Genomes pathways [33], followed by clustering analysis [34], protein-protein interaction (PPI) network prediction [35], and drug-gene interaction [36] to identify the hub genes, key pathways, and potential therapeutic targets. Finally, the obtained gene target network was imported into Cytoscape software (https://cytoscape.org/) for visual editing.
CMAP ANALYSIS AND MOLECULAR DOCKING:
The current version of the Connectivity Map (CMAP) database [37] (build02; www.broadinstitute.org/cmap/) contained more than 7000 expression profiles representing treatments from 1309 compounds. It was an in-silico drug screening approach based on gene expression profiles that had recently been manifested as a prospective strategy for drug repositioning.
Molecular docking was used to verify the targeting association between the compounds and proteins, and the Sybyl X-2.0 [38] was applied to calculate the docking scores represented the binding affinities and obtain detailed information about the relationships between novel drugs and targets.
DATA ANALYSIS:
All data were plotted using GraphPad Prism 7.0. Data were expressed as mean±standard error of the mean (SEM), and the statistical software SPSS24.0 was used for statistical analysis. The
Results
EFFECTS OF AC ON THE HEART RATES AND SV-BA DISTANCE:
After zebrafish embryos exposed to AC, we found that heart rates of larvae increased. As shown in Table 1, compared with the control group, AC 15 mg/L significantly resulted in faster heart rates at 72 hpf (* P<0.001) (Table 1). Over time, zebrafish were still alive while ventricular arrest or irregular heartbeats appeared. The blood enters the atrium via the SV and leaves the ventricle via the BA. In the normal scenario, the atrium and ventricle overlap from the side view. However, if abnormal development appears, the position of the atrium and ventricle changes and the distance between the SV and BA also changes accordingly. AC 15 mg/L reliably increased SV-BA distance relative to controls. (# P<0.001) (Table 1).
MORPHOLOGICAL TOXICITY:
Cardiac structure defects were induced in zebrafish embryos by AC exposure. The toxicity of AC to the cardiac structure mainly includes larger pericardial edema area and blood cell accumulation in the sinus veins and the dorsal aorta. Moreover, the heart structures in AC-treated embryos were elongated and stretched. Because of the less overlap of the atrium and ventricle in the AC-treated group, the SV-BA distance became longer than the looped and shaped hearts in the Control group (Figure 1).
DIFFERENTIAL EXPRESSION ANALYSIS:
Raw data obtained by Illumina HiSeq sequencing were filtered, and clean reads were compared to the reference sequence. DEGs were screened out by differential expression analysis according to gene expression in different sample groups. A total of 1621 DEGs were obtained, among which 696 genes were upregulated and 925 genes were downregulated (Figure 2A). The obtained DEGs were further transferred into the DAVID database finally to get 1380 DEGs in official gene symbols.
The differences in gene expression levels in 2 comparison samples were rapidly examined by the volcano plot, which was a type of scatter plot commonly used to display the results of a differential gene expression analysis. The differential expression between the AC 15 mg/L group and the Control group in the volcano plot are shown in Figure 2B.
GO AND KEGG ENRICHMENT ANALYSIS OF DEGS:
GO was a major bioinformatics initiative used to unify the gene attributes representation across all species. It contains a large amount of experimentally verified and predicted associations between genes and biological terms. The GO annotation system which contains 3 main branches, namely, biological process (BP), molecular function (MF), and cellular component (CC). The top 20 items of our GO enrichment analysis are shown in Figure 2C–2E. GO analysis revealed that AC-induced cardiotoxicity was correlated to a significant enrichment of genes mainly involved the regulation of the cell cycle phase transition in BP; DNA replication and DNA-dependent ATPase activity in MF; and nuclear chromatin and chromosomal region in CC. The most important finding was the enrichment annotation involved in BPs related to the regulation of cell cycle and DNA replication.
Like the GO enrichment principle, KEGG pathway significance enrichment can determine cellular behavior, important biochemical and metabolic pathways, and signal transduction pathways involved in DEGs. A scatter diagram (Figure 2F) is a graphical presentation of our KEGG enrichment analysis results. The top 5 of them were Cell-cycle, DNA replication, p53 signaling pathway, Cellular senescence, and Nucleotide excision repair. The top one item of GO and that of KEGG analysis were both associated with the cell cycle-related biological processes and signaling pathways.
SCREENING OF HUB THERAPEUTIC TARGETS AND KEY SIGNAL PATHWAYS:
The most significant therapeutic targets were selected among the DEGs. Above all, DEGs were input into the STRING database (http://string-db.org/) to obtain the PPI network which revealed the interactions of genes at the protein level. Next, CytoNCA [39], a Cytoscape plugin, was used to obtain clusters of core PPI networks by analyzing the topological properties and calculating betweenness centrality (BC) and degree centrality (DC). With the limiting conditions DC >61 and BC >50, a core PPI network with 27 genes was obtained as depicted in Figure 3A.
Crucially, the drug-gene interaction network of DEGs was screened via the DGIdb database (http://www.dgidb.org/) aiming to identify druggable targets. A total of 136 genes were identified to be druggable molecular targets for AC-induced cardiotoxicity. The core PPI network and drug-gene interaction network were intersected to get the hub genes in AC-induced cardiotoxicity. A total of 10 intersection genes were generated including CDK1, PLK1, AURKB, PCNA, CCNA2, TOP2A, FEN1, TYMS, POLA1, RRM1 (Table 2). Evidently, the aforementioned analysis illustrated that CDK1 was identified as the hub gene target having the highest protein interaction level (DC=192, BC=148) and drug ability (Figure 3B).
To further explore the key signal pathways, 136 druggable genes were used in GO and KEGG enrichment analysis. The results demonstrated that these genes were enriched into 8 signaling pathways (count ≥5) and the vital GO item was Cell-cycle DNA replication. Compared with the GO and KEGG analysis of all DEGs in the previous step, Cell-cycle and DNA replication signaling pathway were identified as key signal pathways of AC cardiotoxicity. (Figure 3C).
VALIDATION OF THE EXPRESSION OF CDK1 IN THE GEO DATABASE:
Myocardial infarction eventually leads to myocardial cell necrosis, which is one of the most severe manifestations coronary artery disease, and a leading cause of death among coronary artery disease patients, so it was particularly important to initiate repair and protection of ischemic myocardium [40]. Controlling the heart rate is one of the key points to reduce the infarct size [41]. The present study demonstrated that zebrafish heart injury via AC exposure was similar to the performance of myocardial infarction. Therefore, we used 3 methods (cloud and local) and 3 types of samples (mouse tissue, human tissue, and single cell) to verify the regulation CDK1 in AC cardiotoxicity and myocardial infarction (Table 3).
Firstly, Biojupies (http://biojupies.cloud) that is a web automated generation application in the cloud was used to obtain the gene expression table, using the method described earlier [42]. The dataset GSE65705 was retrieved using myocardial infarction and human tissue as key search words.
The dataset GSE71906 that is the gene expression matrix of myocardial infarction heart tissue in mouse (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE71906) was screened from the GEO database and then we performed differential gene expression analysis based on the R limma package [43] to further validate the expression of CDK1. The results suggested that in myocardial infarction heart tissue of mouse and human, CDK1 was downregulated, while the degree was lower than that of zebrafish heart injury.
Furthermore, single-cell sequencing, aiming at individual cells genome, was different from ordinary sequencing. Consequently, results of ordinary sequencing would inevitably be affected by different cell heterogeneity, while single-cell sequencing could better help us understand the differences between cells. We screened human myocardial injury single-cell sequencing dataset GSE95140 (
CANDIDATE ANTI-CARDIOTOXICITY COMPOUNDS VIA CMAP ANALYSIS:
136 druggable genes were divided into upregulated and downregulated genes and then uploaded to CMAP to determine potential drugs for cardiotoxicity. The top 10 most significant potential compounds were phenoxybenzamine, parthenolide, tanespimycin, trichostatin A, thioridazine, heptaminol, adiphenine, tranexamic acid, vorinostat, and LY-294002 (Table 4). The chemical structures of these compounds were also downloaded (Figure 4A) from the ZINC database (http://zinc15.docking.org). Among these small compounds, previous studies have proven that heptaminol [44,45] and tranexamic acid [46] can be used in cardiovascular diseases, while the others have the effects of anti-tumor or anti-inflammatory, etc.
CORRELATION BETWEEN CDK1 AND COMPOUNDS VIA MOLECULAR DOCKING:
A molecular docking study was performed to verify the association between the molecules and targets. Using the SYBYL X 2.0 software, the top 10 compounds identified via CMAP analysis were docked with the hub gene CDK1 to obtain docking scores (Table 5) including Total-score and C-score; C-score >3 was considered high binding affinity [47]. Among the top 10 compounds, trichostatin A (Figure 4B) and heptaminol (Figure 4C) had the highest binding affinity to CDK1, which was mutually confirmed by previous studies [48]. Furthermore, heptaminol has been proven to treat cardiovascular system problems such as anti-hypotensive action, right heart failure, and cardiogenic edema [49,50], therefore it was selected as the key drug.
Overall, through the aforementioned comprehensive analysis, GO enrichment analysis results suggested that high-enrichment biological process was the regulation of cell cycle phase transition; Then KEGG enrichment analysis demonstrated that the key signal pathway was the cell-cycle signal pathway. Besides, the hub gene screening results illustrated that CDK1 was the crucial therapeutic target. Besides preliminary verification of the expression of CDK1 was performed in the GEO database. Eventually, to demonstrate the molecular mechanism and therapeutic targets of AC-induced cardiotoxicity more intuitively, a new Hub Genes-Key Pathways interaction network was constructed as shown in Figure 5. Based on this system frame, the hub genes and key signaling pathways of AC-induced cardiotoxicity in zebrafish were clearly labeled.
Discussion
The safety evaluation of chemical drugs, biological agents and especially herbs has always been one of the most crucial issues in pharmacological research. The innovative, well-designed, and systematic safety studies should be carried out step by step to achieve a comprehensive exploration of morphology-molecular mechanisms-gene targets in drug toxicity. The continuous advancement of high-throughput sequencing, bioinformatics analysis, and network pharmacology [51,52] provide new research ideas for pharmacology and toxicology.
With the widespread clinical application of AC, the studies on its toxicity become increasingly urgent. Consistent with previous studies [53], our study, based on the established zebrafish models, demonstrated that cardiotoxicity was the main feature of AC poisoning including atrioventricular arrhythmia, conduction block, and heart arrest. Then, the increase in heart rate caused myocardium overloaded. Besides, it also resulted in myocardial damage, pericardial edema, atria-ventricles dilating, and led to functional and organic damage on the zebrafish heart.
Previous studies revealed that the ion channel, neurotransmitter, energy metabolism, and cell damage of cardiomyocytes may be the mechanism of AC-induced cardiotoxicity [54–56]. However, there was little direct evidence for potential therapeutic targets and biomarkers of AC-induced cardiotoxicity. In the present study, we provided mechanistic insight into previous findings that CDK1 was a promising molecular target and cell-cycle was the most critical biological process and signaling pathway for AC-induced cardiotoxicity using RNA-seq and bioinformatics analysis.
The cell cycle is an ordered event in which a cell replicates its genome, growth, and division. It is regulated by multiple proteins, including cyclins, cyclin kinases, and cyclin kinase inhibitors. Cell cycle is also the basis for life growth, development, and reproduction [57], which includes G1, S, G2, and M phases. CDK1, as a cell-cycle regulator, is crucial in the transition from G1 to S and G2 to M phase [58]. Previous studies have demonstrated that CDK1 and cell-cycle signaling pathways play an important role in arrhythmias and myocardial damage regardless, in patient, mammalian, or zebrafish research. For example, a clinical study suggested that propranolol used to control heart rates could upregulate the cell-cycle related genes in the heart [59]. Another
In a study of cardiomyocyte injury, the activation of CDK1 promoted the re-intervention of cardiomyocyte cell-cycle to enhance cell survival, which induced angiogenesis and reduced the size of infarction. Therefore, the cardiac structure and function were jointly improved in mice with myocardial infarction [61]. A recent investigation of the cardiomyocyte illustrated that overexpression of CDK1 activated proliferation of postmitotic cardiomyocytes. It also enhanced cardiac function and increased cardiomyocyte proliferation after myocardial infarction [62]. Overall, in light of our study results and previous research, we can conclude that AC inhibited the cell-cycle signaling pathway by downregulating the expression of CDK1 which resulted in myocardial injury and cardiac rhythm changes. The regulation of cell-cycle related genes stimulated adult cardiomyocyte proliferation and cardiac regeneration. It supported the notion that cell-cycle reactivation used carefully may be effective as a therapeutic strategy for cardiac repair [63].
Pharmacogenomics encompassed potentially clinically actionable gene-drug associations, which may greatly accelerate the process of drug development. According to our screening and prediction through the CMAP database and molecular docking, heptaminol was one of the key compounds for anti-AC cardiotoxicity. Meanwhile, our study also found that cardiac injury induced by AC exposure in zebrafish mainly included pericardial edema and atrial enlargement. Heptaminol was widely recognized for the treatment for right heart failure, coronary heart disease, and cardiogenic edema, which can improve atrioventricular node excitability, excite the conduction system, and restore its normal function [44,45,64].
Unexpectedly, we found that AC could result in the downregulation of PLK1, CCNB1, etc. However, these genes were demonstrated to be overexpressed in cancer samples in previous studies [65–67]. Furthermore, part of the top 10 small molecules selected via our CMAP analysis possessed an anti-tumor effect. Therefore, our data supported the hypothesis that AC had potential anticancer activity, which provides references for herbal drugs to treat cancer in the future.
Even if the current study has proposed drug candidates for augmenting the present management of AC cardiotoxicity, several limitations need to be resolved for future improvements. First, although zebrafish have certain advantages in the study of heart disease, future experimental studies will need to be expanded to include rats, cells, and other models to explore the pharmacological and toxicological mechanisms of AC in different species. Second, the potential drugs identified in the current study need to be validated in further
Conclusions
In conclusion, we investigated the manifestation and analyzed the possible mechanism of AC-induced cardiotoxicity in zebrafish. Furthermore, the AC cardiotoxicity-related genes identified via RNA-seq and bioinformatics analysis will attract more related research. The candidate drugs screened through the CMAP database and molecule docking may offer innovative treatment perspectives of drug toxicity and provide a systematic bioinformatics method to illustrate the molecular mechanisms of toxicology.
Figures
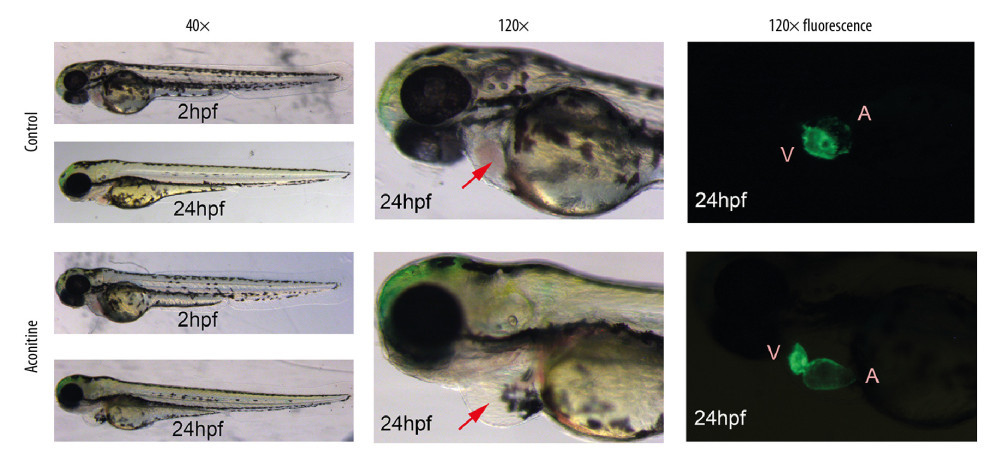 Figure 1. Morphology and heart fluorescence images of zebrafish embryos on the effects of aconitine (AC). V – ventricle; A – atria. AC group versus control group, significant pericardial edema and blood cell accumulation in AC-exposed zebrafish hearts (marked with red arrow).
Figure 1. Morphology and heart fluorescence images of zebrafish embryos on the effects of aconitine (AC). V – ventricle; A – atria. AC group versus control group, significant pericardial edema and blood cell accumulation in AC-exposed zebrafish hearts (marked with red arrow). 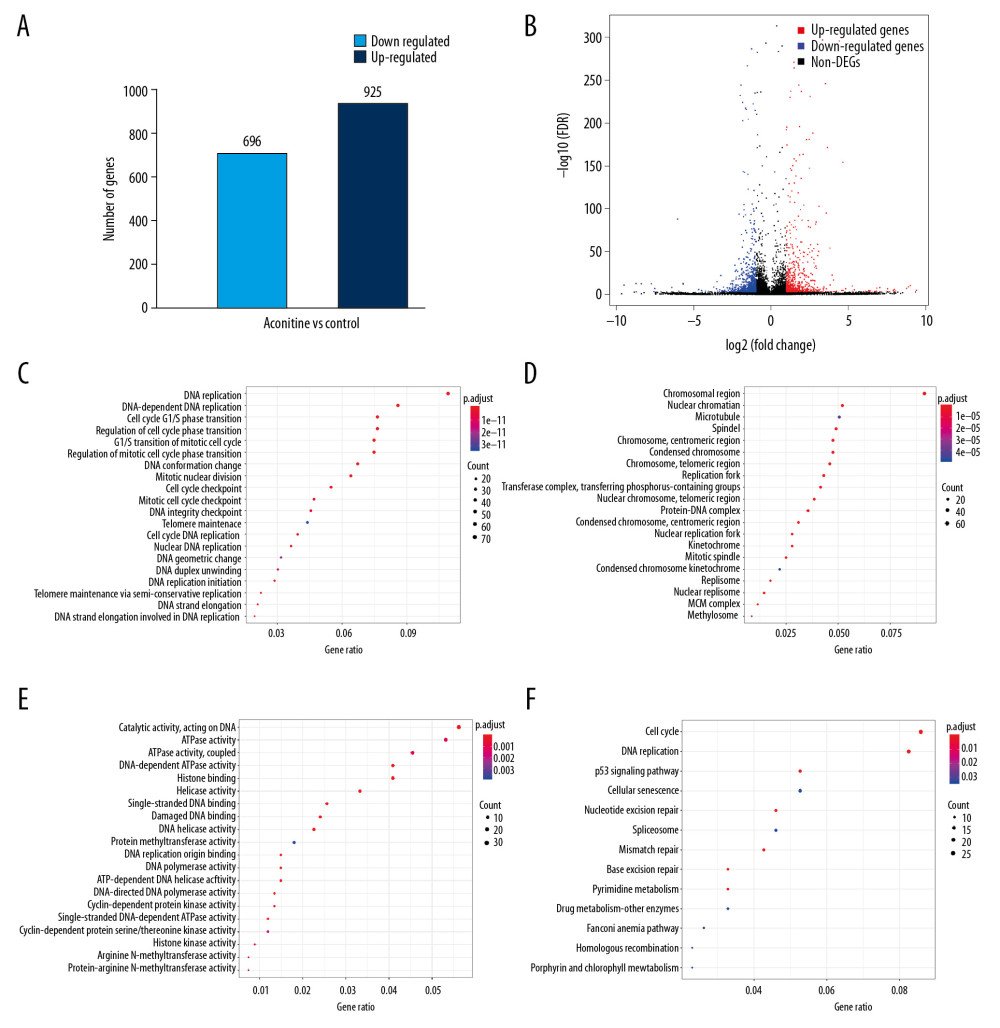 Figure 2. (A) Statistical chart of the number of DEGs. The horizontal axis represents a comparison of different samples, the vertical axis represents the number of DEGs, red represents upregulation, blue represents downregulation. (B) Volcano plot of DEGs. The figure contains an interactive scatter plot that displays the log2-fold changes and statistical significance of each gene calculated by performing a differential gene expression analysis. Each dot in the volcano plot of DEGs represents a gene. Red points indicate significantly upregulated genes, blue points indicate downregulated genes, and the black dots represent non-differentially expressed genes. The axes display the significance versus fold-change estimated by the differential expression analysis. The results of the GO and KEGG enrichment analysis of all DEGs. (C) Biological process. (D) Cellular component. (E) Molecular function. (F) KEGG pathway. The size of the point represents the number of genes enriched, and the depth of the color represents the size of the P-value. The X-axis is the name of the items arranged according to the degree of enrichment, and the Y-axis is the size of the gene ratio. DEGs – differentially expressed genes; GO – Gene Ontology; KEGG – Kyoto Encyclopedia of Genes and Genomes.
Figure 2. (A) Statistical chart of the number of DEGs. The horizontal axis represents a comparison of different samples, the vertical axis represents the number of DEGs, red represents upregulation, blue represents downregulation. (B) Volcano plot of DEGs. The figure contains an interactive scatter plot that displays the log2-fold changes and statistical significance of each gene calculated by performing a differential gene expression analysis. Each dot in the volcano plot of DEGs represents a gene. Red points indicate significantly upregulated genes, blue points indicate downregulated genes, and the black dots represent non-differentially expressed genes. The axes display the significance versus fold-change estimated by the differential expression analysis. The results of the GO and KEGG enrichment analysis of all DEGs. (C) Biological process. (D) Cellular component. (E) Molecular function. (F) KEGG pathway. The size of the point represents the number of genes enriched, and the depth of the color represents the size of the P-value. The X-axis is the name of the items arranged according to the degree of enrichment, and the Y-axis is the size of the gene ratio. DEGs – differentially expressed genes; GO – Gene Ontology; KEGG – Kyoto Encyclopedia of Genes and Genomes. 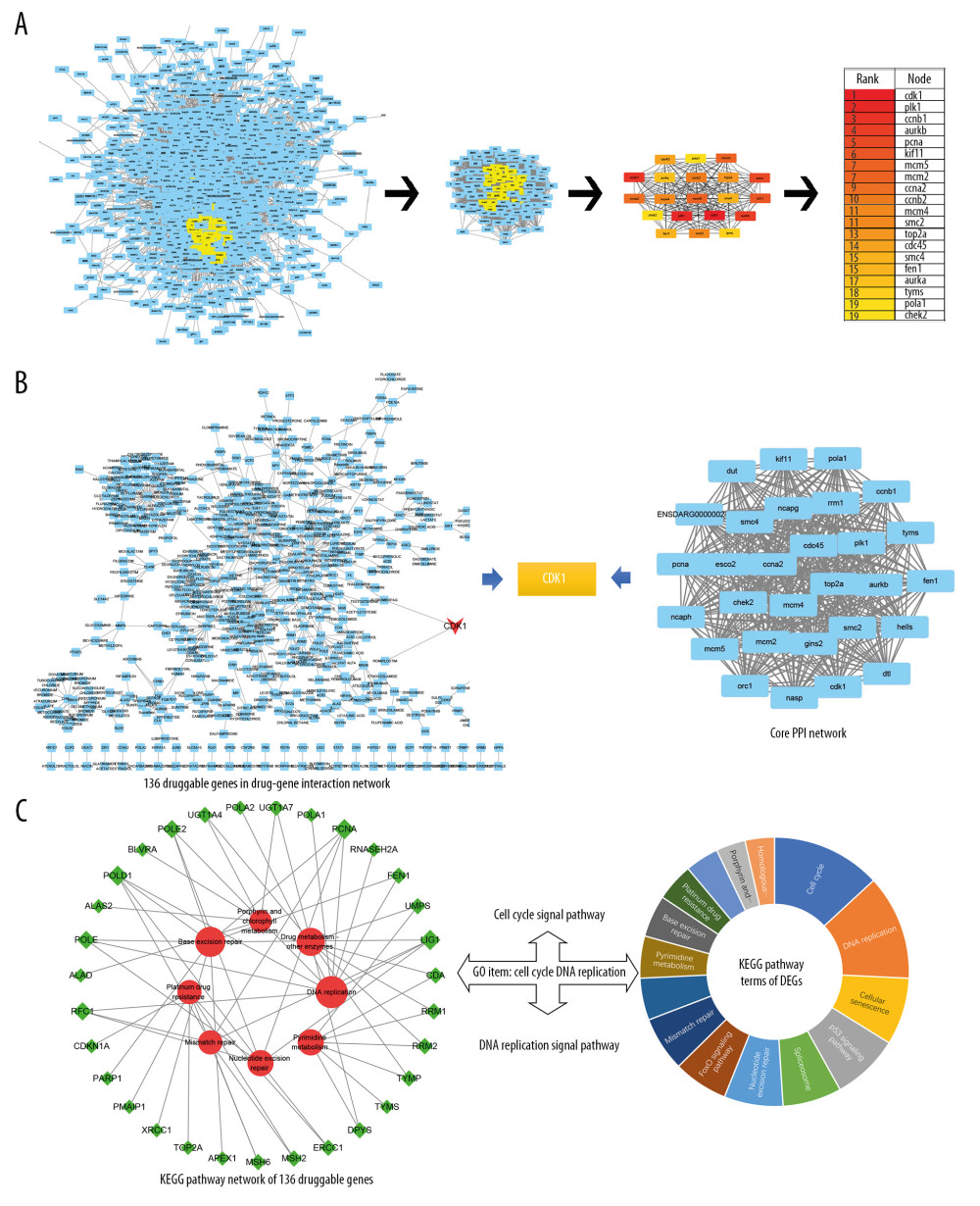 Figure 3. (A) Core PPI network and hub genes identified by CytoNCA and Cytoscape. More important nodes received higher quantitative values within the network. The list of the sequence of the top 20 genes. The shade of the color indicates the degree of the gene. (B) Screen hub therapeutic target gene. drug-gene interaction network (left) and core PPI network (right). (C) Key signal pathways. A network of KEGG-druggable genes (left) and top 20 KEGG items of DEGs (right). PPI – protein-protein interaction; KEGG – Kyoto Encyclopedia of Genes and Genomes; DEGs – differentially expressed genes.
Figure 3. (A) Core PPI network and hub genes identified by CytoNCA and Cytoscape. More important nodes received higher quantitative values within the network. The list of the sequence of the top 20 genes. The shade of the color indicates the degree of the gene. (B) Screen hub therapeutic target gene. drug-gene interaction network (left) and core PPI network (right). (C) Key signal pathways. A network of KEGG-druggable genes (left) and top 20 KEGG items of DEGs (right). PPI – protein-protein interaction; KEGG – Kyoto Encyclopedia of Genes and Genomes; DEGs – differentially expressed genes. 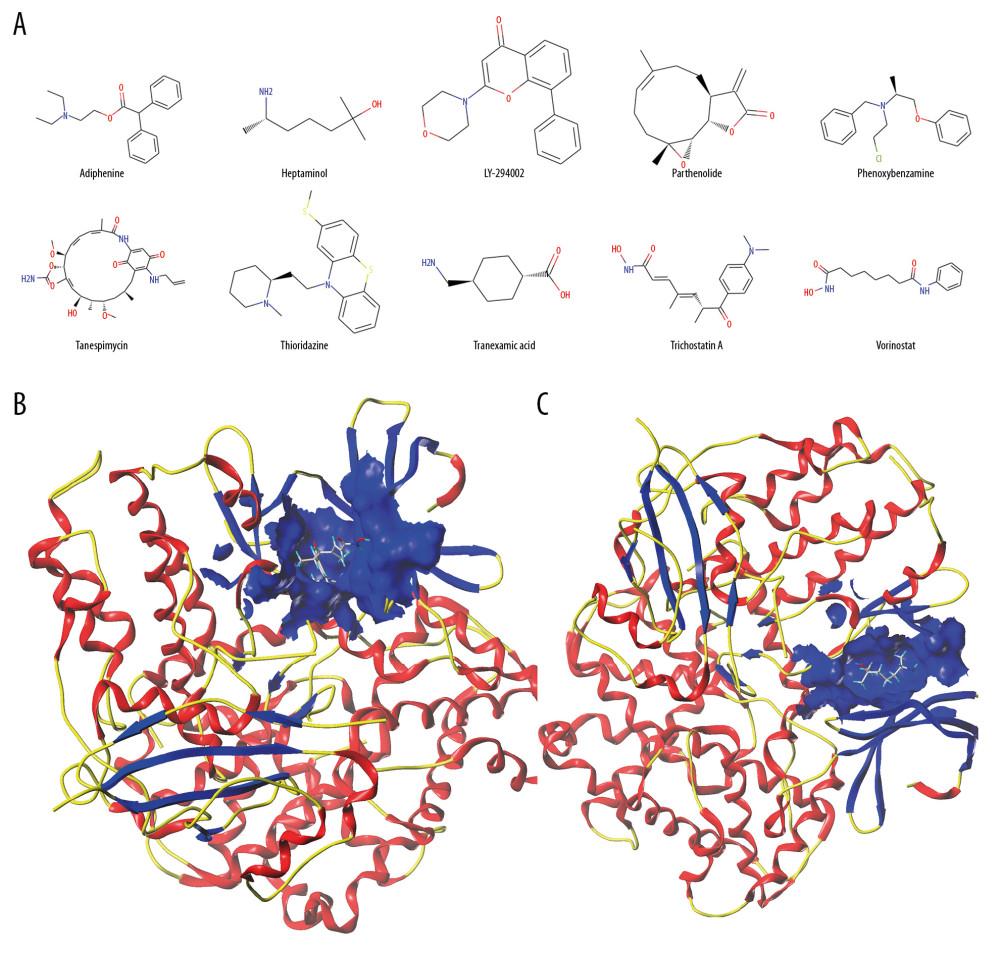 Figure 4. (A) The chemical structures of top 10 compounds via CMAP analysis. (B) Binding of trichostatin A and CDK1. (C) Binding of heptaminol and CDK1. CMAP – connectivity map; CDK1 – cyclin-dependent kinase-1.
Figure 4. (A) The chemical structures of top 10 compounds via CMAP analysis. (B) Binding of trichostatin A and CDK1. (C) Binding of heptaminol and CDK1. CMAP – connectivity map; CDK1 – cyclin-dependent kinase-1. 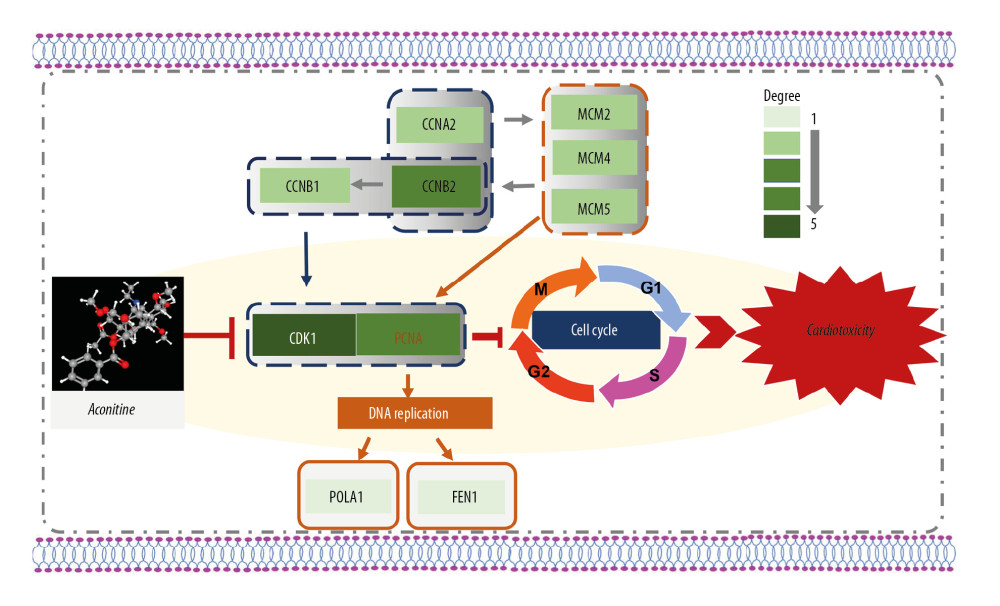 Figure 5. A new hub genes-key pathway network. A systematic understanding of the mechanism of AC cardiotoxicity. The depth of the color represents the number of genes enriched in the pathway. CDK1 and cell-cycle signal pathway play a key role in aconitine-induced cardiotoxicity. CDK1 – cyclin-dependent kinase-1; AC – aconitine.
Figure 5. A new hub genes-key pathway network. A systematic understanding of the mechanism of AC cardiotoxicity. The depth of the color represents the number of genes enriched in the pathway. CDK1 and cell-cycle signal pathway play a key role in aconitine-induced cardiotoxicity. CDK1 – cyclin-dependent kinase-1; AC – aconitine. Tables
Table 1. Heart rates and SV-BA of zebrafish in each group at 72 hpf (n=16).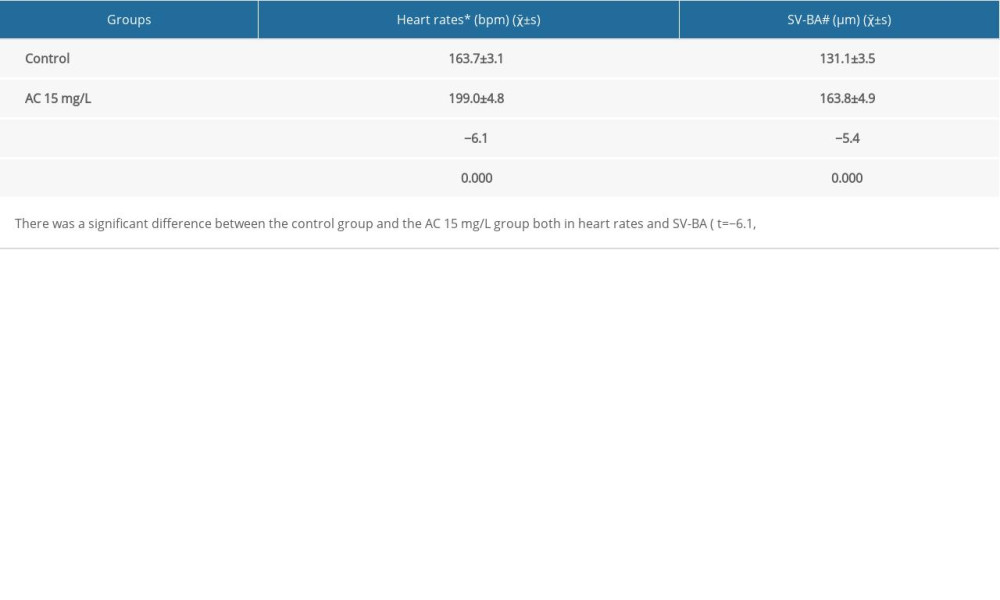 Table 2. Topological parameters of the targets.
Table 2. Topological parameters of the targets.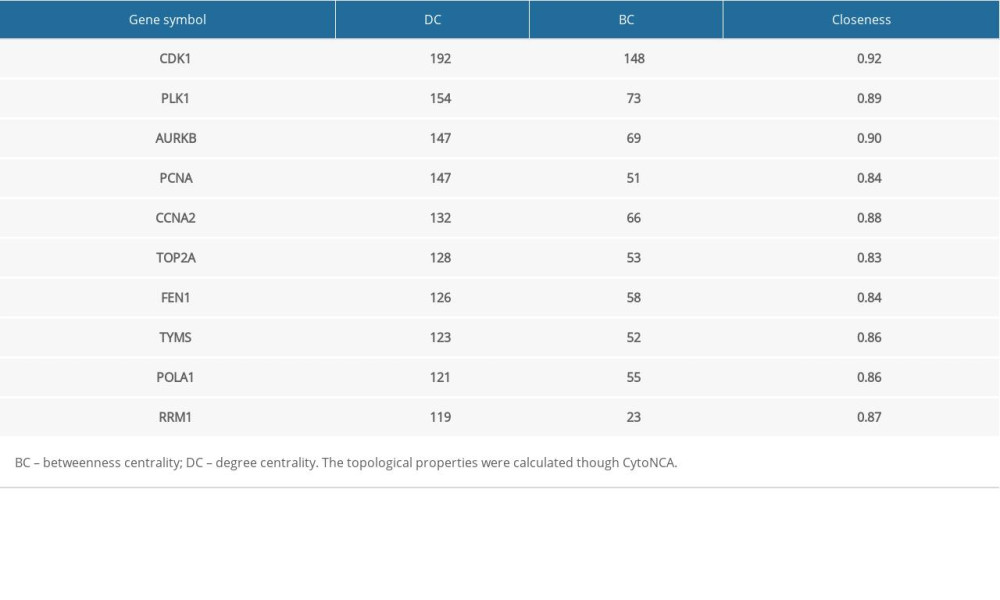 Table 3. Validation the expression of CDK1 using different method and tissues.
Table 3. Validation the expression of CDK1 using different method and tissues.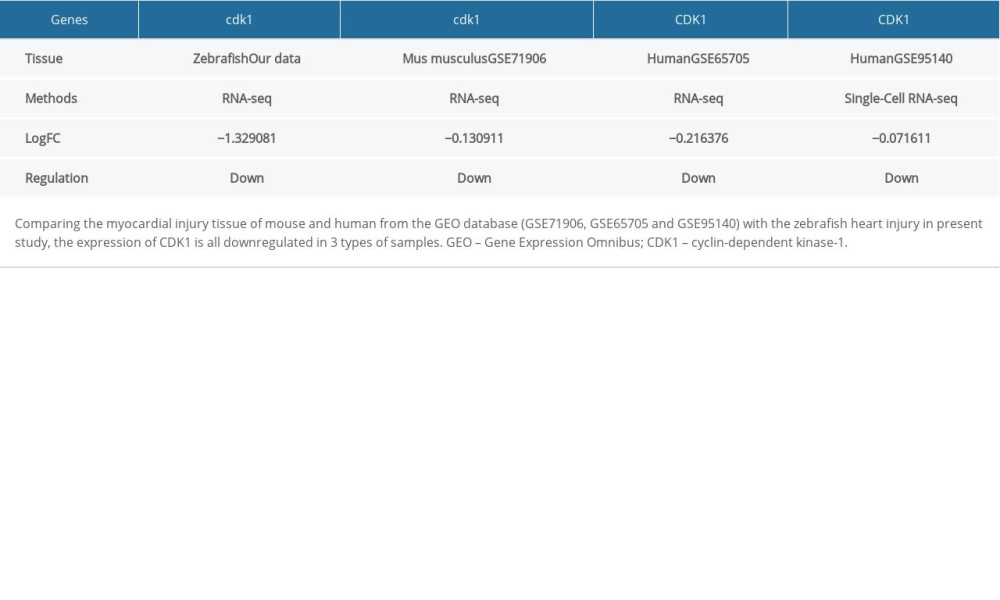 Table 4. Top 10 most significant compounds based on CMAP for AC-induced cardiotoxicity.
Table 4. Top 10 most significant compounds based on CMAP for AC-induced cardiotoxicity.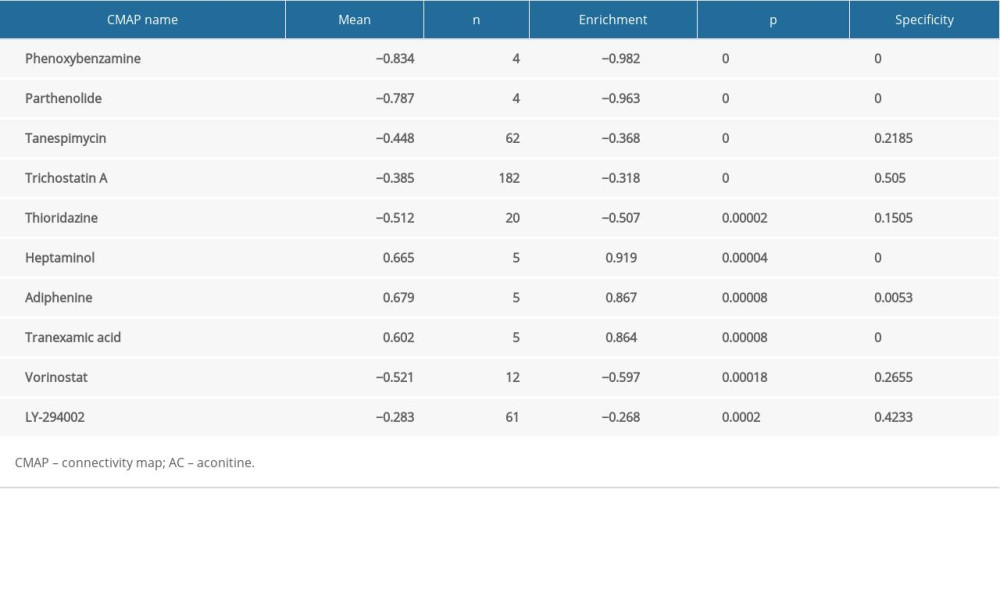 Table 5. Molecular docking results via Sybyl-X 2.0.
Table 5. Molecular docking results via Sybyl-X 2.0.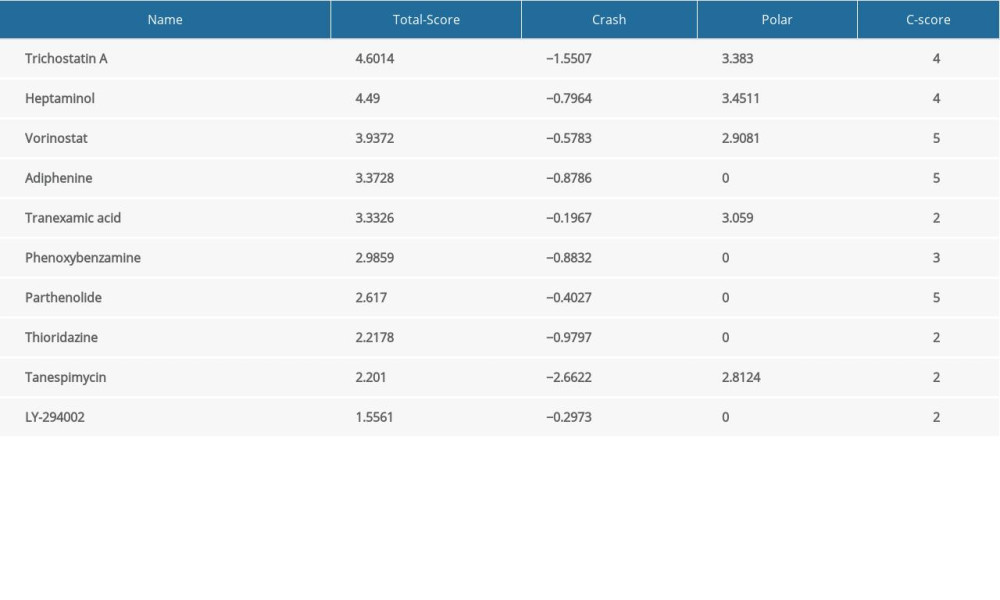
References
1. Gao T, Bi H, Ma S, Lu J: Nat Prod Commun, 2010; 5; 447-55
2. Yang Z, Lu ZQ, Zhang YJ, Looking for agonists of beta2 adrenergic receptor from Fuzi and Chuanwu by virtual screening and dual-luciferase reporter assay: J Asian Nat Prod Res, 2016; 18; 550-61
3. Guo Q, Xia H, Shi G, Zhang T, Shi J: Org Lett, 2018; 20; 816-19
4. Ji BL, Xia LP, Zhou FX, Aconitine induces cell apoptosis in human pancreatic cancer via NF-kappaB signaling pathway: Eur Rev Med Pharmacol Sci, 2016; 20; 4955-64
5. Li TF, Gong N, Wang YX, Ester hydrolysis differentially reduces aconitine-induced anti-hypersensitivity and acute neurotoxicity: Involvement of spinal microglial dynorphin expression and implications for aconitum processing: Front Pharmacol, 2016; 7; 367
6. Li H, Liu L, Zhu S, Liu Q, Case reports of aconite poisoning in mainland China from 2004 to 2015: A retrospective analysis: J Forensic Leg Med, 2016; 42; 68-73
7. Nyirimigabo E, Xu Y, Li Y, A review on phytochemistry, pharmacology and toxicology studies of Aconitum: J Pharm Pharmacol, 2015; 67; 1-19
8. Zhou G, Tang L, Zhou X: J Ethnopharmacol, 2015; 160; 173-93
9. Chan TY, Aconitum alkaloid poisoning related to the culinary uses of aconite roots: Toxins (Basel), 2014; 6; 2605-11
10. Coulson JM, Caparrotta TM, Thompson JP, The management of ventricular dysrhythmia in aconite poisoning: Clin Toxicol (Phila), 2017; 55; 313-21
11. Zong X, Yan X, Wu JL: J Nat Prod, 2019; 82; 980-89
12. Li M, Xie X, Chen H, Aconitine induces cardiotoxicity through regulation of calcium signaling pathway in zebrafish embryos and in H9c2 cells: J Appl Toxicol, 2020 [Epub ahead of print]
13. Ma LQ, Yu Y, Chen H, Sweroside alleviated aconitine-induced cardiac toxicity in H9c2 cardiomyoblast cell line: Front Pharmacol, 2018; 9; 1138
14. Peng F, Zhang N, Wang C, Aconitine induces cardiomyocyte damage by mitigating BNIP3-dependent mitophagy and the TNFα-NLRP3 signalling axis: Cell Prolif, 2020; 53; e12701
15. Rocke J, Lees J, Packham I, Chico T, The zebrafish as a novel tool for cardiovascular drug discovery: Recent Pat Cardiovasc Drug Discov, 2009; 4; 1-5
16. Bakkers J, Zebrafish as a model to study cardiac development and human cardiac disease: Cardiovasc Res, 2011; 91; 279-88
17. Takada N, Omae M, Sagawa F, Re-evaluating the functional landscape of the cardiovascular system during development: Biol Open, 2017; 6; 1756-70
18. Conesa A, Madrigal P, Tarazona S, Erratum to: A survey of best practices for RNA-seq data analysis: Genome Biol, 2016; 17; 181
19. Wong VK-W, Law BY-K, Yao X-J, Advanced research technology for discovery of new effective compounds from Chinese herbal medicine and their molecular targets: Pharmacol Res, 2016; 111; 546-55
20. Li GM, Zhang CL, Rui RP, Bioinformatics analysis of common differential genes of coronary artery disease and ischemic cardiomyopathy: Eur Rev Med Pharmacol Sci, 2018; 22; 3553-69
21. Cotto KC, Wagner AH, Feng Y-Y, DGIdb 3.0: A redesign and expansion of the drug-gene interaction database: Nucleic Acids Res, 2018; 46; D1068-73
22. Lamb J, The connectivity map: A new tool for biomedical research: Nat Rev Cancer, 2007; 7; 54-60
23. Wang Z, Dai Z, Luo Z, Zuo C, Identification of pyrvinium, an anthelmintic drug, as a novel anti-adipogenic compound based on the gene expression microarray and connectivity map: Molecules, 2019; 24(13) pii: E2391
24. Lin P, Zhong X, Wang X, Survival analysis of genome-wide profiles coupled with Connectivity Map database mining to identify potential therapeutic targets for cholangiocarcinoma: Oncol Rep, 2018; 40; 3189-98
25. Zhu F-X, He Y-C, Zhang J-Y, Using prognosis-related gene expression signature and connectivity map for personalized drug repositioning in multiple myeloma: Med Sci Monit, 2019; 25; 3247-55
26. Westerfield M, 1995, University of Oregon press
27. Fang F, Jie Z, Linzhong Y, Jiabo L, Preliminary investigation of cardiac toxicity to zebrafish embryo by Aconitine: Pharm Clin Chin Materia Medica, 2012; 2; 32-34
28. Sinha JN, Shamsi MA, Kohli RP, Bhargava KP, Centrogenic cardiac arrhythmia induced by aconitine: A new “model” for screening of anti-arrhythmic drugs: Jpn J Pharmacol, 1971; 21; 699-706
29. Kessler M, Berger IM, Just S, Rottbauer W, Loss of dihydrolipoyl succinyltransferase (DLST) leads to reduced resting heart rate in the zebrafish: Basic Res Cardiol, 2015; 110; 14
30. Du Z, Wang G, Gao S, Wang Z, Aryl organophosphate flame retardants induced cardiotoxicity during zebrafish embryogenesis: By disturbing expression of the transcriptional regulators: Aquat Toxicol, 2015; 161; 25-32
31. Han Y, Zhang JP, Qian JQ, Hu CQ: J Appl Toxicol, 2015; 35; 241-52
32. Chen K, Liu J, Liu S, Methyltransferase SETD2-mediated methylation of STAT1 Is critical for interferon antiviral activity: Cell, 2017; 170; 492-506.e14
33. Harris MA, Clark J, Ireland A, The Gene Ontology (GO) database and informatics resource: Nucleic Acids Res, 2004; 32; D258-61
34. Huang da W, Sherman BT, Lempicki RA, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources: Nat Protoc, 2009; 4; 44-57
35. Szklarczyk D, Gable AL, Lyon D, STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets: Nucleic Acids Res, 2019; 47; D607-13
36. Wagner AH, Coffman AC, Ainscough BJ, DGIdb 2.0: Mining clinically relevant drug-gene interactions: Nucleic Acids Res, 2016; 44; D1036-44
37. Lamb J, Crawford ED, Peck D, The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease: Science, 2006; 313; 1929-35
38. Jain AN, Surflex: Fully automatic flexible molecular docking using a molecular similarity-based search engine: J Med Chem, 2003; 46; 499-511
39. Tang Y, Li M, Wang J, CytoNCA: A cytoscape plugin for centrality analysis and evaluation of protein interaction networks: Biosystems, 2015; 127; 67-72
40. Thygesen K, Alpert JS, Jaffe AS, Fourth universal definition of myocardial infarction (2018): Circulation, 2018; 138; e618-51
41. Reed GW, Rossi JE, Cannon CP, Acute myocardial infarction: Lancet, 2017; 389; 197-210
42. Torre D, Lachmann A, Ma’ayan A, BioJupies: Automated generation of interactive notebooks for RNA-Seq data analysis in the cloud: Cell Syst, 2018; 7; 556-61.e3
43. Mehmood A, Laiho A, Venäläinen MS, Systematic evaluation of differential splicing tools for RNA-seq studies: Brief Bioinform, 2019 [Epub ahead of print]
44. Peineau N, Mongo KG, Le Guennec JY, Alteration of the L-type calcium current in guinea-pig single ventricular myocytes by heptaminol hydrochloride: Br J Pharmacol, 1992; 107; 104-8
45. Edno-Mcheik L, Gaulier JM, Combourieu I, Heptaminol interferes in the AxSYM FPIA amphetamine/methamphetamine II assay: Clin Chem, 2001; 47; 1499-500
46. Myles PS, Smith JA, Forbes A, Tranexamic acid in patients undergoing coronary-artery surgery: N Engl J Med, 2017; 376; 136-48
47. Joshi SD, Dixit SR, Kirankumar MN, Synthesis, antimycobacterial screening and ligand-based molecular docking studies on novel pyrrole derivatives bearing pyrazoline, isoxazole and phenyl thiourea moieties: Eur J Med Chem, 2016; 107; 133-52
48. Zhu FX, He YC, Zhang JY, Using prognosis-related gene expression signature and connectivity map for personalized drug repositioning in multiple myeloma: Med Sci Monit, 2019; 25; 3247-55
49. Peineau N, Mongo KG, Le Guennec JY, Alteration of the L-type calcium current in guinea-pig single ventricular myocytes by heptaminol hydrochloride: Br J Pharmacol, 1992; 107; 104-8
50. Milon D, Allain H, Reymann JM, Randomized double-blind trial of injectable heptaminol for controlling spontaneous or bromocriptine-induced orthostatic hypotension in parkinsonians: Fundam Clin Pharmacol, 1990; 4; 695-705
51. Afshari CA, Hamadeh HK, Bushel PR, The evolution of bioinformatics in toxicology: Advancing toxicogenomics: Toxicol Sci, 2011; 120(Suppl 1); S225-37
52. Li M, Wang J, Fu L: Med Sci Monit, 2020; 26; e920376
53. Liu S, Li F, Li Y, A review of traditional and current methods used to potentially reduce toxicity of aconitum roots in traditional Chinese medicine: J Ethnopharmacol, 2017; 207; 237-50
54. Gao X, Zhang X, Hu J, Aconitine induces apoptosis in H9c2 cardiac cells via mitochondria mediated pathway: Mol Med Rep, 2018; 17; 284-92
55. Luo JX, Zhang Y, Hu XY: J Ethnopharmacol, 2016; 183; 187-92
56. Zhou YH, Piao XM, Liu X, Arrhythmogenesis toxicity of aconitine is related to intracellular ca(2+) signals: Int J Med Sci, 2013; 10; 1242-49
57. Puente BN, Kimura W, Muralidhar SA, The oxygen-rich postnatal environment induces cardiomyocyte cell-cycle arrest through DNA damage response: Cell, 2014; 157; 565-79
58. Fededa JP, Gerlich DW, Molecular control of animal cell cytokinesis: Nat Cell Biol, 2012; 14; 440-47
59. Musumeci M, Maccari S, Sestili P, Propranolol enhances cell cycle-related gene expression in pressure overloaded hearts: Br J Pharmacol, 2011; 164; 1917-28
60. Sai C, Yunhan J, Zhao J, Cyclin dependent kinase 1 (CDK1) activates cardiac fibroblasts via directly phosphorylating paxillin at Ser244: Int Heart J, 2019; 60; 374-83
61. Borden A, Kurian J, Nickoloff E, Transient introduction of miR-294 in the heart promotes cardiomyocyte cell cycle reentry after injury: Circ Res, 2019; 125; 14-25
62. Mohamed TMA, Ang YS, Radzinsky E, Regulation of cell cycle to stimulate adult cardiomyocyte proliferation and cardiac regeneration: Cell, 2018; 173; 104-16.e12
63. Hsieh PC, Kamp TJ, To be young at heart: Cell Stem Cell, 2018; 22; 475-76
64. Berthiau F, Garnier D, Argibay JA, Decrease in internal H+ and positive inotropic effect of heptaminol hydrochloride: a 31P N.M.R. spectroscopy study in rat isolated heart: Br J Pharmacol, 1989; 98; 1233-40
65. de Cárcer G, Venkateswaran SV, Salgueiro L, Plk1 overexpression induces chromosomal instability and suppresses tumor development: Nat Commun, 2018; 9; 3012
66. Hassan QN, Alinari L, Byrd JC, PLK1: A promising and previously unexplored target in double-hit lymphoma: J Clin Invest, 2018; 128; 5206-8
67. Xie B, Wang S, Jiang N, Li JJ, Cyclin B1/CDK1-regulated mitochondrial bioenergetics in cell cycle progression and tumor resistance: Cancer Lett, 2019; 443; 56-66
Figures
 Figure 1. Morphology and heart fluorescence images of zebrafish embryos on the effects of aconitine (AC). V – ventricle; A – atria. AC group versus control group, significant pericardial edema and blood cell accumulation in AC-exposed zebrafish hearts (marked with red arrow).
Figure 1. Morphology and heart fluorescence images of zebrafish embryos on the effects of aconitine (AC). V – ventricle; A – atria. AC group versus control group, significant pericardial edema and blood cell accumulation in AC-exposed zebrafish hearts (marked with red arrow). Figure 2. (A) Statistical chart of the number of DEGs. The horizontal axis represents a comparison of different samples, the vertical axis represents the number of DEGs, red represents upregulation, blue represents downregulation. (B) Volcano plot of DEGs. The figure contains an interactive scatter plot that displays the log2-fold changes and statistical significance of each gene calculated by performing a differential gene expression analysis. Each dot in the volcano plot of DEGs represents a gene. Red points indicate significantly upregulated genes, blue points indicate downregulated genes, and the black dots represent non-differentially expressed genes. The axes display the significance versus fold-change estimated by the differential expression analysis. The results of the GO and KEGG enrichment analysis of all DEGs. (C) Biological process. (D) Cellular component. (E) Molecular function. (F) KEGG pathway. The size of the point represents the number of genes enriched, and the depth of the color represents the size of the P-value. The X-axis is the name of the items arranged according to the degree of enrichment, and the Y-axis is the size of the gene ratio. DEGs – differentially expressed genes; GO – Gene Ontology; KEGG – Kyoto Encyclopedia of Genes and Genomes.
Figure 2. (A) Statistical chart of the number of DEGs. The horizontal axis represents a comparison of different samples, the vertical axis represents the number of DEGs, red represents upregulation, blue represents downregulation. (B) Volcano plot of DEGs. The figure contains an interactive scatter plot that displays the log2-fold changes and statistical significance of each gene calculated by performing a differential gene expression analysis. Each dot in the volcano plot of DEGs represents a gene. Red points indicate significantly upregulated genes, blue points indicate downregulated genes, and the black dots represent non-differentially expressed genes. The axes display the significance versus fold-change estimated by the differential expression analysis. The results of the GO and KEGG enrichment analysis of all DEGs. (C) Biological process. (D) Cellular component. (E) Molecular function. (F) KEGG pathway. The size of the point represents the number of genes enriched, and the depth of the color represents the size of the P-value. The X-axis is the name of the items arranged according to the degree of enrichment, and the Y-axis is the size of the gene ratio. DEGs – differentially expressed genes; GO – Gene Ontology; KEGG – Kyoto Encyclopedia of Genes and Genomes. Figure 3. (A) Core PPI network and hub genes identified by CytoNCA and Cytoscape. More important nodes received higher quantitative values within the network. The list of the sequence of the top 20 genes. The shade of the color indicates the degree of the gene. (B) Screen hub therapeutic target gene. drug-gene interaction network (left) and core PPI network (right). (C) Key signal pathways. A network of KEGG-druggable genes (left) and top 20 KEGG items of DEGs (right). PPI – protein-protein interaction; KEGG – Kyoto Encyclopedia of Genes and Genomes; DEGs – differentially expressed genes.
Figure 3. (A) Core PPI network and hub genes identified by CytoNCA and Cytoscape. More important nodes received higher quantitative values within the network. The list of the sequence of the top 20 genes. The shade of the color indicates the degree of the gene. (B) Screen hub therapeutic target gene. drug-gene interaction network (left) and core PPI network (right). (C) Key signal pathways. A network of KEGG-druggable genes (left) and top 20 KEGG items of DEGs (right). PPI – protein-protein interaction; KEGG – Kyoto Encyclopedia of Genes and Genomes; DEGs – differentially expressed genes. Figure 4. (A) The chemical structures of top 10 compounds via CMAP analysis. (B) Binding of trichostatin A and CDK1. (C) Binding of heptaminol and CDK1. CMAP – connectivity map; CDK1 – cyclin-dependent kinase-1.
Figure 4. (A) The chemical structures of top 10 compounds via CMAP analysis. (B) Binding of trichostatin A and CDK1. (C) Binding of heptaminol and CDK1. CMAP – connectivity map; CDK1 – cyclin-dependent kinase-1. Figure 5. A new hub genes-key pathway network. A systematic understanding of the mechanism of AC cardiotoxicity. The depth of the color represents the number of genes enriched in the pathway. CDK1 and cell-cycle signal pathway play a key role in aconitine-induced cardiotoxicity. CDK1 – cyclin-dependent kinase-1; AC – aconitine.
Figure 5. A new hub genes-key pathway network. A systematic understanding of the mechanism of AC cardiotoxicity. The depth of the color represents the number of genes enriched in the pathway. CDK1 and cell-cycle signal pathway play a key role in aconitine-induced cardiotoxicity. CDK1 – cyclin-dependent kinase-1; AC – aconitine. Tables
 Table 1. Heart rates and SV-BA of zebrafish in each group at 72 hpf (n=16).
Table 1. Heart rates and SV-BA of zebrafish in each group at 72 hpf (n=16). Table 2. Topological parameters of the targets.
Table 2. Topological parameters of the targets. Table 3. Validation the expression of CDK1 using different method and tissues.
Table 3. Validation the expression of CDK1 using different method and tissues. Table 4. Top 10 most significant compounds based on CMAP for AC-induced cardiotoxicity.
Table 4. Top 10 most significant compounds based on CMAP for AC-induced cardiotoxicity. Table 5. Molecular docking results via Sybyl-X 2.0.
Table 5. Molecular docking results via Sybyl-X 2.0. Table 1. Heart rates and SV-BA of zebrafish in each group at 72 hpf (n=16).
Table 1. Heart rates and SV-BA of zebrafish in each group at 72 hpf (n=16). Table 2. Topological parameters of the targets.
Table 2. Topological parameters of the targets. Table 3. Validation the expression of CDK1 using different method and tissues.
Table 3. Validation the expression of CDK1 using different method and tissues. Table 4. Top 10 most significant compounds based on CMAP for AC-induced cardiotoxicity.
Table 4. Top 10 most significant compounds based on CMAP for AC-induced cardiotoxicity. Table 5. Molecular docking results via Sybyl-X 2.0.
Table 5. Molecular docking results via Sybyl-X 2.0. In Press
06 Mar 2024 : Clinical Research
Comparison of Outcomes between Single-Level and Double-Level Corpectomy in Thoracolumbar Reconstruction: A ...Med Sci Monit In Press; DOI: 10.12659/MSM.943797
21 Mar 2024 : Meta-Analysis
Economic Evaluation of COVID-19 Screening Tests and Surveillance Strategies in Low-Income, Middle-Income, a...Med Sci Monit In Press; DOI: 10.12659/MSM.943863
10 Apr 2024 : Clinical Research
Predicting Acute Cardiovascular Complications in COVID-19: Insights from a Specialized Cardiac Referral Dep...Med Sci Monit In Press; DOI: 10.12659/MSM.942612
06 Mar 2024 : Clinical Research
Enhanced Surgical Outcomes of Popliteal Cyst Excision: A Retrospective Study Comparing Arthroscopic Debride...Med Sci Monit In Press; DOI: 10.12659/MSM.941102
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952








