02 September 2020: Database Analysis
A Novel Risk Model Based on Autophagy Pathway Related Genes for Survival Prediction in Lung Adenocarcinoma
Fan Zhang1ABCG, Suzhen Xie1DE, Zhenyu Zhang1C, Huanhuan Zhao1E, Zijun Zhao1F, Haiying Sun2G, Jiao Zheng1A*DOI: 10.12659/MSM.924710
Med Sci Monit 2020; 26:e924710
Abstract
BACKGROUND: Autophagy has a principal role in mediating tumor cell metabolism. However, the role of autophagy-pathway-related genes (APRGs) as prognostic markers remains obscure in lung adenocarcinoma (LUAD). More potential prognostic biomarkers are needed to deepen our understanding to explore the prognostic role of APRGs in LUAD.
MATERIAL AND METHODS: We used The Cancer Genome Atlas (TCGA) database to identify differentially expressed APRGs. Cox proportional hazard regression was used to identify prognostic APRGs, and then a risk model was constructed. The efficacy of the risk model was confirmed using a testing group. Lastly, we explored mutational signatures of prognostic of APRGs. T-tests were used to analyze all the expression patterns of genes by SPSS 19.0.
RESULTS: Using TCGA database, 5 differently expressed APRGs were identified in LUAD patients, and functional enrichment analyze of the genes that were closely associated with the survival status in LUAD patients. Cox proportional hazard regression was facilitated to identify 9 APRGs (CCR2, LAMP1, RELA, ATG12, ATG9A, NCKAP1, ATG10, DNAJB9, and MBTPS2). Multivariate Cox proportional hazards regression analyses further identified 5 key prognostic APRGs (CCR2, LAMP1, RELA, ATG12, and MBTPS2) that were closely related to the survival status in LUAD. Then the prognostic scores based on the 5 genes as independent prognostic indicators were constructed for overall survival (OS) of LUAD patients; area under the curve (AUC) values >0.70 (all P<0.05). The efficacy of prognostic scores was confirmed by data from the testing group and showed significant differences between the low-risk and the high-risk groups for OS (P<0.05).
CONCLUSIONS: The risk model based on the construction of 5 APRGs can predict the prognosis of patients with LUAD, which may potentially predict prognostic signatures for LUAD.
Keywords: Autophagy-Related Proteins, Databases, Genetic, Mutation
Background
Lung carcinoma is one of the most frequent malignancies and a leading cause of cancer-related death while lung adenocarcinoma (LUAD) is considered the most common histological subtype with a sudden onset and unexpected 5-year survival rate [1,2]. Therapeutic options for LUAD vary considerably, representing a complicated obstacle to disease management [3]. Though effective targeted medicines have been rapidly invented, it is still the most lethal cancer in the world [4]. Unfortunately, very few biomarkers have been discovered to efficiently predict prognosis of LUAD patients [5]. Thus, exploring accurate markers for predicting the survival of LUAD is an urgent scientific problem.
Autophagy refers to a catabolic process that vesicles of the double-membrane structure are encapsulate damaged or denatured necrotic organelles in the cytoplasm to form autophagosomes, and then these autophagosomes fused with lysosomes are digested and degraded. These processes are regulated by specific genes that could adjust inflammation, immune response, oxidative stress, and so on [6,7]. Autophagy has been shown to have dual effects on cancer [8]. It mostly depends on the circumstance of cells whether autophagy plays a suppressed role or promoted role [9]. With the increasing research on the pathogenesis of autophagy, the study of autophagy in cancer has correspondingly increased. Autophagy-pathway-related genes (APRGs) which can be used as the prognostic signatures are required for autophagy [10,11]. However, the role of APRGs in the prognostic assessment in LUAD is still unknown. Thus, systematic studies for more accurate prognostic markers as well as novel and reliable therapeutic targets will be useful for assessment of tumor progression, metastasis, and recurrence risk of LUAD. Although it is well known that autophagy plays a crucial role in lung cancer, the role of autophagy in LUAD remains unclear and limited.
In this research, we searched and downloaded APRGs and analyzed RNA sequence data as well as clinicopathological characteristics of LUAD patients from The Cancer Genome Atlas (TCGA), then we performed a functional analysis of differentially expressed genes. To identify prognostic APRGs, Cox proportional hazard regression model was established to study the association between the expression of these obtained genes and the survival of LUAD patients. The prognostic score was used as an assessment for the patient’s risk score (Figure 1). These findings are expected to effectively monitor autophagy and predict the prognosis of patients with LUAD.
Material and Methods
DATA MINING:
In our analysis, 234 APRGs were downloaded from the Human Autophagy Database (HADb,
FUNCTIONAL OF DIFFERENTIALLY EXPRESSED APRGS:
To estimate the underlying mechanism of differentially expressed APRGs, the downloaded data was analyzed by the R package edgeR program, with a value of |log2(Fold Change)| >1 and
IDENTIFICATION OF PROGNOSTIC BIOMARKERS FOR LUAD:
To identify the prognostic genes and validate the efficacy of risk score, we studied 458 patients with LUAD for whom we could obtain entire overall survival (OS) information (Table 1). These patients were randomly divided into 2 groups, a training group (n=230) and a testing group (n=228). The data from the training group was used to construct the Cox regression model, while data from the testing group was used to confirm the steadiness of model. Lasso regression analysis was used to get a more refined model. Next, these prognostic genes were analyzed by univariate Cox regression analysis with the expression data standardized into [log2 (data+1)]. Ultimately, the prognosis related APRGs with a P value <0.05 by univariate analysis were chosen for multivariate Cox analysis. Patients of the training group were divided into a high-risk group and a low-risk group according to their median risk score. The accuracy of the risk score was evaluated using the survival receiver operator characteristic (ROC).
CONSTRUCTION OF AN INDIVIDUALIZED PROGNOSTIC INDEX:
To further analyzed the relationship between the risk score and clinical parameters, multivariate Cox regression analysis was performed according to gender, age, pathological stage, and the risk score obtained from the prognostic model as covariates.
MUTATIONAL SIGNATURES OF PROGNOSTIC APRGS IN LUAD:
To analyze the mutational frequency of the prognostic genes that are related to OS in LUAD patients, a total of 564 samples from TCGA dataset were included in this research.
STATISTICAL ANALYSIS:
All the data in our study were analyzed by SPSS 19.0 (IBM SPSS) and R 3.2.2. Multivariate and univariate Cox analyses analyzed the independent prognostic value by using survival package of R. T-tests were used to analyze the expression patterns of genes in LUAD and the clinicopathological parameters. A
Results
IDENTIFICATION OF DIFFERENTIALLY EXPRESSED APRGS IN LUAD:
By searching the data of LUAD in TCGA, 551 cases of the RNASeq data (54 normal issues and 497 tumor issues) as well as prognosis data of 486 LUAD patients were obtained. Totally 234 genes related to autophagy were extracted via HADb. By using edgeR software, 5 differentially expressed APRGs between tumor tissues and normal tissues were identified for further study (P<0.001, Figure 2). 4 genes were remarkably higher expressed than in tumor issues while 1 gene was remarkably lower expressed (P<0.001).
FUNCTIONAL ANNOTATION OF DIFFERENTIALLY EXPRESSED APRGS:
To explore the biological function and the pathways of the differentially expressed APRGs, we further performed GO and KEGG analysis of all the differently expressed APRGs in LUAD. The most enriched GO item for biological processes (BP) was “positive regulation of tyrosine phosphorylation of STAT protein”; changes in molecular function (MF) were mainly enriched in “ubiquitin-protein ligase binding” (Figure 3). However, KEGG pathway enrichment analysis showed there was no statistically significant pathway.
IDENTIFICATION OF PROGNOSTIC APRGS IN THE TRAINING GROUP:
The univariate Cox analysis and Lasso method initially identified a total of 9 APRGs (CCR2, LAMP1, RELA, ATG12, ATG9A, NCKAP1, ATG10, DNAJB9, and MBTPS2) that had a close relationship between gene expression and OS in the training group (Figure 4A, 4B). The significantly different variables were identified in the multivariate analyses. After that, CCR2, LAMP1, RELA, ATG12, and MBTPS2 were selected as independent prognostic indicators for OS of LUAD. These optimal APRGs with prognostic values for LUAD were used to establish a risk score model, which was based on the expression of the risk genes for the OS model. The risk model for OS was as follows (Table 2): risk score=(−0.2807×expression value of CCR2)+(0.0008×expression value of LAMP1)+(0.0466×expression value of RELA)+ (0.2662×expression value of ATG12)+(0.1513×expression value of MBTPS2). According to the median risk, patients in the training group were divided into a high-risk group (n=115) and a low-risk group (n=115). Kaplan-Meier analysis showed that patients with low-risk prognosis had significantly better survival than patients with high-risk prognosis (P=4.67e-05, Figure 4C). Besides, the number of LUAD patients in the training group was ranked by the risk scores (Figure 5A, 5C, 5E). Our analyses showed that the 5-year OS rate for the high-risk group was 23.1% (95% CI=12.6–42.1%), whereas the low-risk group was 56.9% (95% CI=41.8%–77.4%). The AUC value for the ROC curve was 0.711 for 5 years, which implied that the 5 APRGs prognosis risk assessment model was credible and effective (Figure 4E).
VALIDATION OF PROGNOSTIC APRGS IN THE TESTING GROUP:
To validate the prediction accuracy of the prognostic risk model, the testing group served as the validation set, by calculating the risk scores of each patient (Figure 5B, 5D, 5F). We also divided the testing group patients into a high-risk group (n=114) and a low-risk group (n=114) in the OS model. Kaplan-Meier survival curve analysis showed significant differences between the high-risk group and the low-risk group for OS (P=4.79e-04) (Figure 4D). Our analysis showed that 5-year OS rates for the high-risk group and low-risk group were 18.0% (95% CI=9.21–35.3%) and 56.9% (95% CI=41.8–77.4%) respectively, which indicated that patients in the low-risk group also had better OS than patients in the high-risk group. The AUC value for the OS model was 0.703 at 5 years (Figure 4F).
PROGNOSTIC RISK MODELS ARE INDEPENDENTLY ASSOCIATED WITH OS OF LUAD PATIENTS:
We also analyzed the relationship between the risk model and key clinical parameters such as age, gender, stage, and pathological factors (Table 3). In univariate analysis, we found that T-stage (hazard ratio [HR]=1.632, 95% CI: 1.315–2.024, P<0.001), N-stage (HR=1.790, 95% CI: 1.459–2.196, P<0.001), tumor stage (HR=1.654 95% CI: 1.401–1.951, P<0.001), risk score (HR=1.380, 95% CI: 1.283–1.484, P<0.001) were suggested as independent prognostic factors (Figure 6A). Moreover, multivariate analysis suggested that tumor stage (HR=1.930 95% CI: 1.190–3.130, P<0.01) and risk score (HR=1.318, 95% CI: 1.218–1.425, P<0.001) were selected as the significant risk factors for LUAD patients with complementary value (Figure 6B). These results demonstrated that the prognostic model can be independently used to predict OS for LUAD patients. We then assessed the accuracy of the risk score model using the ROC curve analysis, the area under ROC curve (AUC) values were 0.705 and 0.723, for risk score and stage respectively.
MUTATIONAL ANALYSIS OF 5 PROGNOSTIC APRGS INVOLVED IN THE LUAD NETWORK:
The mutational analyses on the 5 prognostic APRGs showed a significant amplification in tumor tissues (Figure 7). 4 out of 5 prognostic genes revealed more than 0.9% genetic alterations in LUAD. Gene LAMP1 had the highest rates of alteration (2.6%) followed by MBTPS2 (1.7%), ATG12 (0.9%), and CCR2 (0.9%). RELA had the lowest rate of alteration (0.4%).
Discussion
LUAD is one of the leading causes of malignant lung tumors and involved high morbidity and mortality rates in 2019 [12]. The inefficient prognosis of LUAD patients leads to drug resistance and metastasis. It is clear that exploring the prognosis of LUAD may affect outcomes of patients with LUAD and will significantly benefit the diagnosis, treatment, and prognosis assessment.
Autophagy serves a pivotal role in maintaining cellular homeostasis and diminishes tumorigenesis [13]. The relationship between autophagic responses and tumor has been widely studied. Previous research has shown that APRGs can mediate the death of tumor cells [14]. Autophagy not only promotes proliferation to favor of tumor cells, but also induces apoptosis of cells. Recent research has demonstrated that ectopic expression of KISS1 downregulates the 2 key modulators of autophagy, ATG5 and ATG7. This funding implicates autophagy is involved in the mechanism of breast cancer metastatic transformation [15]. Current clinical practice evaluation of the outcome of patients who suffer from LUAD mainly depends on TMN stage, which is not precise enough. The prognostic effects of APRGs in LUAD remains to be elucidated [16,17].
During the past decades, deep mining of omics big data became an important assessment for the diagnosis, treatment, and prognosis of cancer. TCGA program provides an easy access to obtain high-quality tumor genomic data with clinical information. Herein, with the assistance of TCGA, we first mined the public data of patients with LUAD and found that 5 APRGs were significant differentially expressed in tumor samples. Then we performed bioinformatics analyses using multiple bioinformatics tools. This showed that the differently expressed APRGs tended to be an important process involved in the evolution of LUAD, providing clues to the underlying mechanisms of LUAD. By performing univariate analyses and multivariate analyses, a list of 5 core prognostic APRGs (CCR2, LAMP1, RELA, ATG12, and MBTPS2) was used to facilitate our risk model, showing their effect on the outcomes of patients with LUAD. Finally, we investigated the correlation of the expression of APRGs and clinic features. All the results demonstrated our risk score may well predict prognosis and provide novel messages for clinical application in LUAD patients. CCR2 studies in cancer research have mostly focused on regulating immune cell recruitment, which can drive fundamental mechanisms such as growth, angiogenesis, and progression of a variety of tumors including lung cancer [18,19]. LAMP1 is an important regulator of maturation of autophagosomes and phagosomes. Directly targeting LAMP1 can modulate autophagy by accelerating autophagosome formation and degradation. Moreover, LAMP1 is involved in inhibition of autophagy-mediated proliferation and metastasis of pancreatic ductal adenocarcinoma [20]. ATG12, a key member of ATG family, has been shown to mediate its functions to conjugate to ATG5 and ATG3 to promote mitochondrial fusion and mitochondrial apoptosis [21]. Sun et al. [22] also found that targeting ATG12 can exert influence on the promotion of autophagy and regulation of EPI sensitivity in breast cancer cells. RELA, a protein subunit of NF-κB, mainly exerts its effects on post-translational modifications, including acetylation, phosphorylation, ubiquitination, and methylation, and has been proven to be implicated in numerous types of tumorigenesis [23]. Previous research has shown that overexpression of RELA can inhibited proliferation and apoptosis rates of lung cancer cells [24]. Interestingly, our results indicated that high expression of RELA in LUAD tissues was associated with poor prognosis. MBTPS2, which exists in the Golgi membrane, is an intramembranous metalloprotease and plays a role in the regulation of cholesterol metabolism or ER stress response [25]. There is ample evidence to show that MBTPS2 mutation contributes to many common diseases, such as diabetic nephropathy [26], ichthyosis follicularis [27], and so on, but there is a lack of research on MBTPS2 in relation to autophagy and cancer progression.
In summary, we identified 5 key prognostic APRGs that could provide a more accurate estimate of OS in LUAD patients. These findings may also provide a multidimensional biomarker strategy that can more effectively and comprehensively message the role of autophagy in LUAD and may hold great promise for enhancing diagnostic accuracy and predicting treatment response. The main limitation of our findings was that our study was conducted and used data already collected from patients in public databases. Nevertheless, further prospective experimental studies are needed to validate prospective clinical trials and the mechanisms of APRGs modulating the initiation and progression of LUAD.
Conclusions
Based on TCGA, we identified 5 prognostic APRGs and developed a risk model to better predict the OS prognosis of LUAD patients, and determined the independent prognosis of LUAD patients, and which could provide accurate novel prognosis predictor for high-risk population screening and provide another promising predictive biomarker for clinical practice in LUAD treatment.
Figures
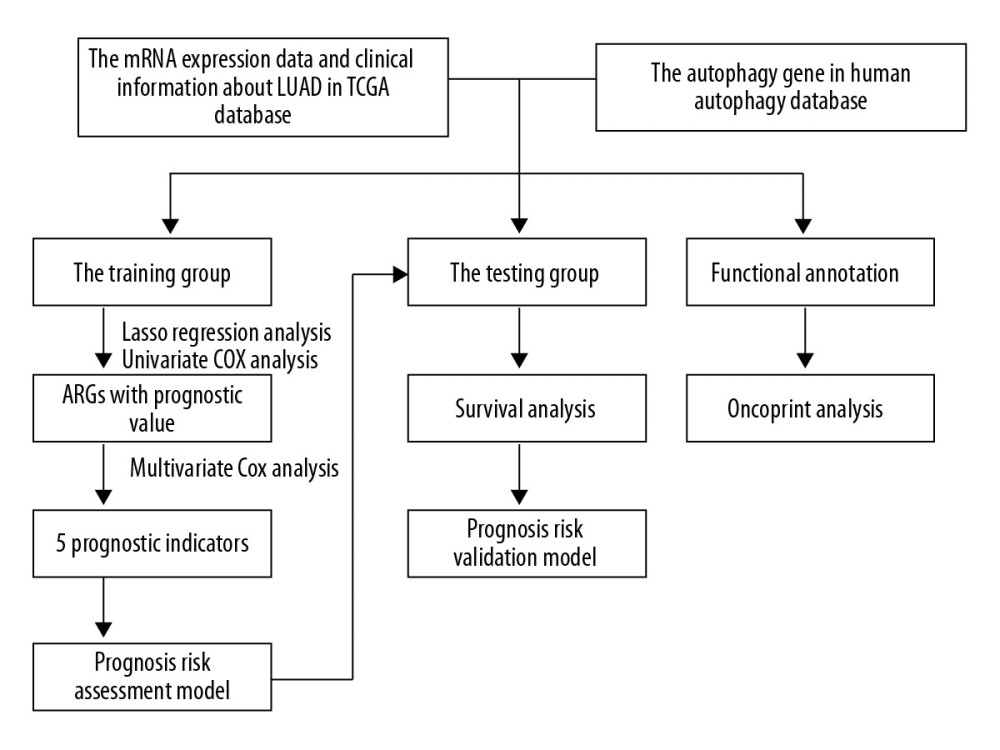 Figure 1. The flowchart of identification of LUAD survival-related autophagy prognostic signature. Abbreviations: LUAD, lung adenocarcinoma.
Figure 1. The flowchart of identification of LUAD survival-related autophagy prognostic signature. Abbreviations: LUAD, lung adenocarcinoma. 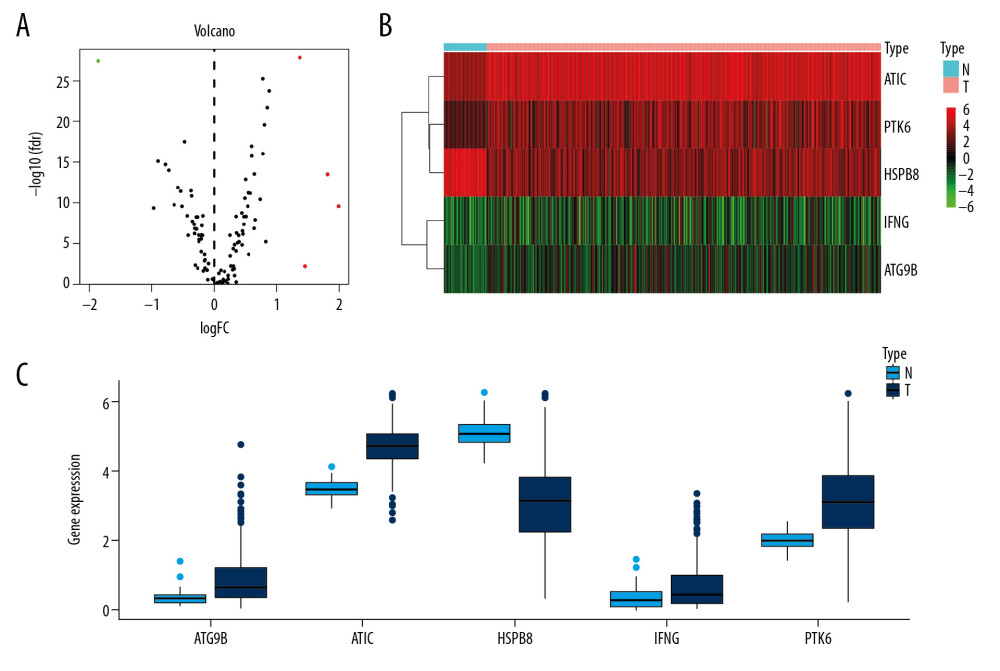 Figure 2. Differentially expressed APRGs between lung carcinoma and normal tissues. (A) The volcano plot of 5 differentially expressed APRGs. The red dots represent the level of high expression and the green dots represent the level of low expression. (B) Heatmap of 5 differently expressed APRGs. The depth of red represents the level of high expression, and the depth of green represents the level of low expression. (C) The boxplot of 5 differentially expressed APRGs. Abbreviations: APRGs, autophagy-pathway-related genes.
Figure 2. Differentially expressed APRGs between lung carcinoma and normal tissues. (A) The volcano plot of 5 differentially expressed APRGs. The red dots represent the level of high expression and the green dots represent the level of low expression. (B) Heatmap of 5 differently expressed APRGs. The depth of red represents the level of high expression, and the depth of green represents the level of low expression. (C) The boxplot of 5 differentially expressed APRGs. Abbreviations: APRGs, autophagy-pathway-related genes. 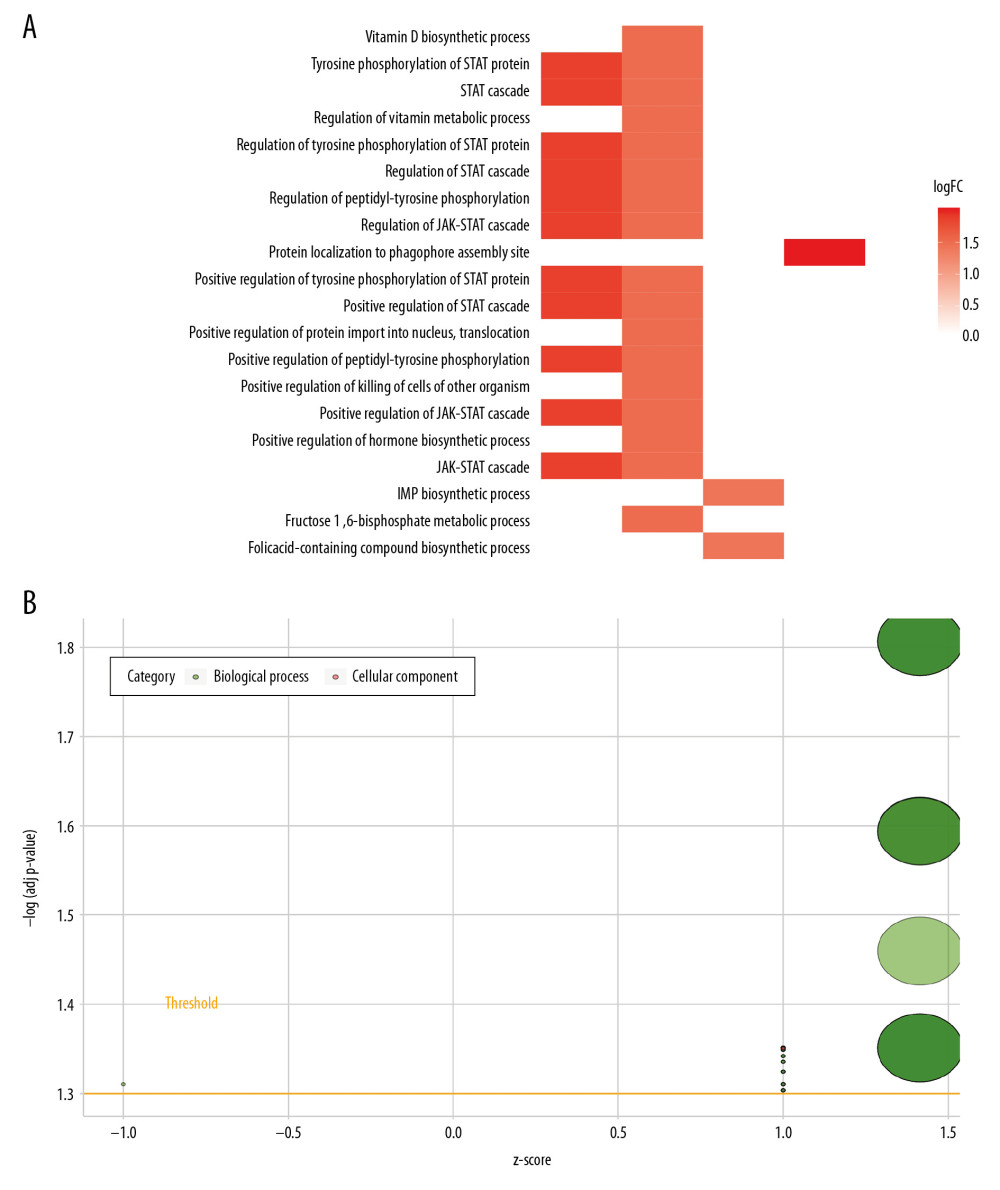 Figure 3. The heat plot (A) and bubble plot (B) of enriched Gene Ontology (GO) terms. The change in color from blue to red represents the increase in the adjusted P-value, and the length of the bar indicates the number of gene enrichment terms.
Figure 3. The heat plot (A) and bubble plot (B) of enriched Gene Ontology (GO) terms. The change in color from blue to red represents the increase in the adjusted P-value, and the length of the bar indicates the number of gene enrichment terms. 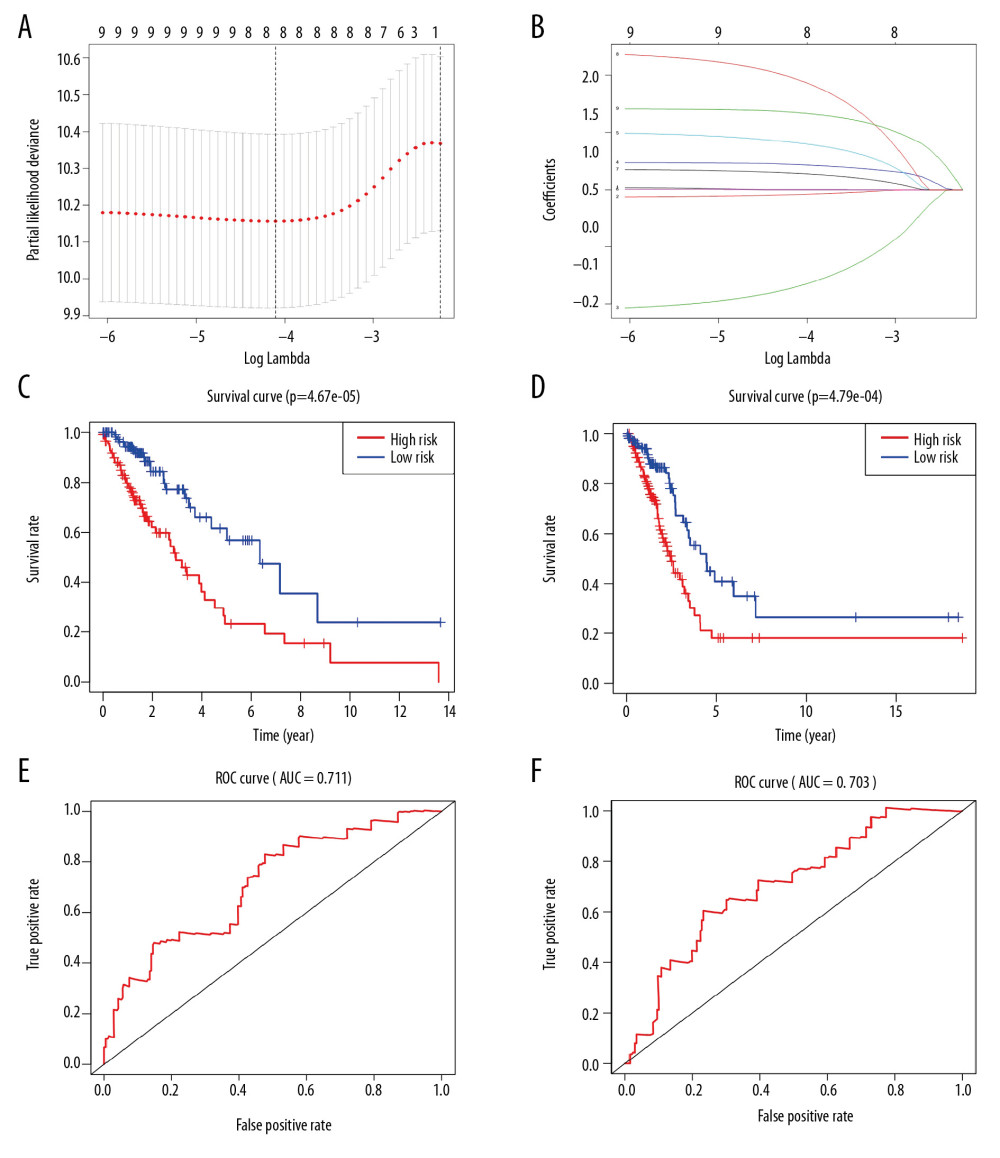 Figure 4. Prognostic index of 5 APRGs. (A) Lasso regression of 5 APRGs using the OS model. (B) Lasso co-efficient profiles of 5 APRGs by optimal lambda. Kaplan-Meier plot represents that patients in the high-risk group (red line) had significantly shorter overall survival time than those in the low-risk group (blue line). (C) Left: training group (D) Right: testing group. Time-dependent ROC curve analysis shows AUC values for OS in LUAD patients. (E) Left: training group. (F) Right: testing group. APRGs – autophagy-pathway-related genes; ROC – receiver-operator characteristic; AUC – area under the curve; OS – overall survival. LUAD – lung adenocarcinoma.
Figure 4. Prognostic index of 5 APRGs. (A) Lasso regression of 5 APRGs using the OS model. (B) Lasso co-efficient profiles of 5 APRGs by optimal lambda. Kaplan-Meier plot represents that patients in the high-risk group (red line) had significantly shorter overall survival time than those in the low-risk group (blue line). (C) Left: training group (D) Right: testing group. Time-dependent ROC curve analysis shows AUC values for OS in LUAD patients. (E) Left: training group. (F) Right: testing group. APRGs – autophagy-pathway-related genes; ROC – receiver-operator characteristic; AUC – area under the curve; OS – overall survival. LUAD – lung adenocarcinoma. 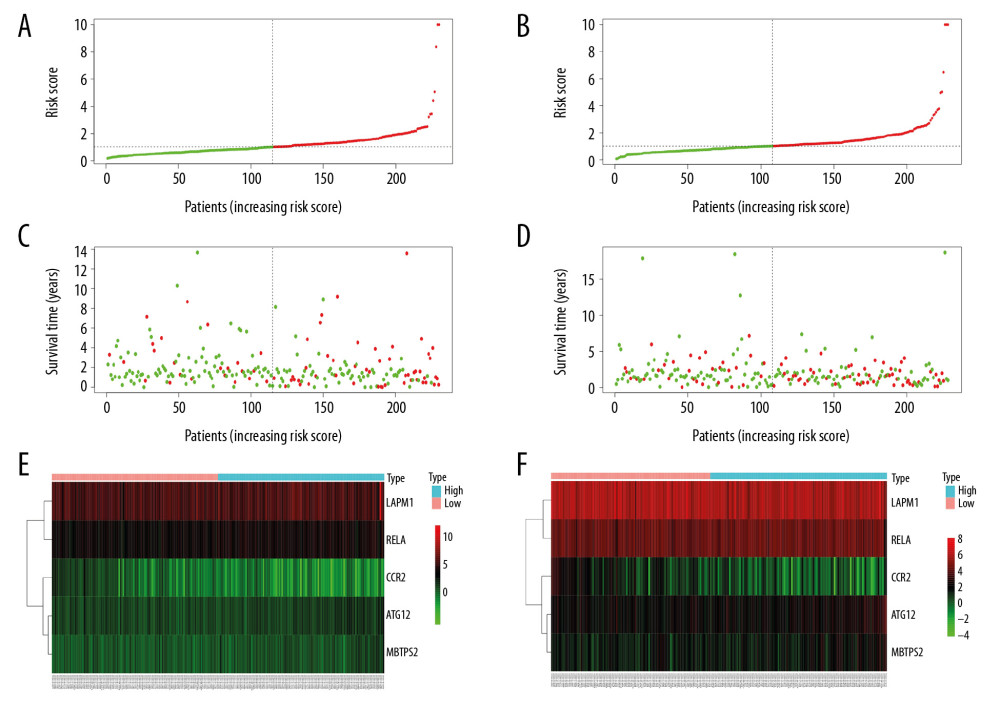 Figure 5. Prognosis of high-risk and low-risk LUAD patients. Risk score distribution of low-risk (green) and high-risk (red) in LUAD patients in (A) training group and (B) testing group. Scatter plot of survival status of LUAD patients in (C) training group and (D) testing group. Red dots (dead); green dots (alive). Expression of risk genes in the high-risk (blue) and low-risk (pink) of the OS model in (E) training group and (F) testing group. LUAD – lung adenocarcinoma; OS – overall survival.
Figure 5. Prognosis of high-risk and low-risk LUAD patients. Risk score distribution of low-risk (green) and high-risk (red) in LUAD patients in (A) training group and (B) testing group. Scatter plot of survival status of LUAD patients in (C) training group and (D) testing group. Red dots (dead); green dots (alive). Expression of risk genes in the high-risk (blue) and low-risk (pink) of the OS model in (E) training group and (F) testing group. LUAD – lung adenocarcinoma; OS – overall survival. 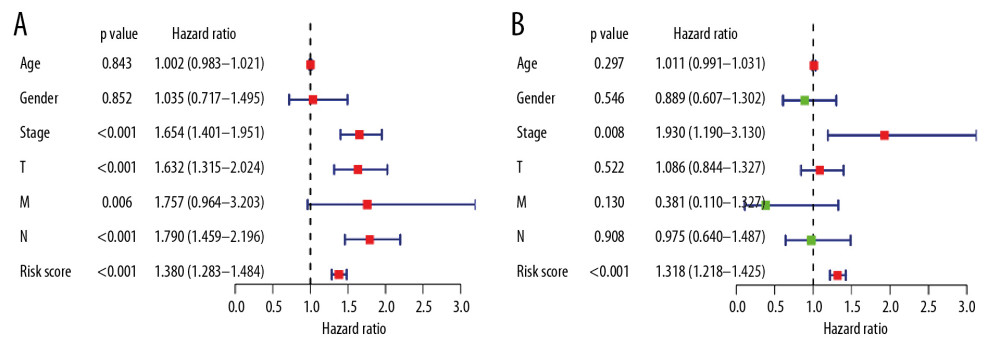 Figure 6. (A) Univariate and (B) multivariate analyses of OS in LUAD patients. OS – overall survival; LUAD – lung adenocarcinoma.
Figure 6. (A) Univariate and (B) multivariate analyses of OS in LUAD patients. OS – overall survival; LUAD – lung adenocarcinoma. 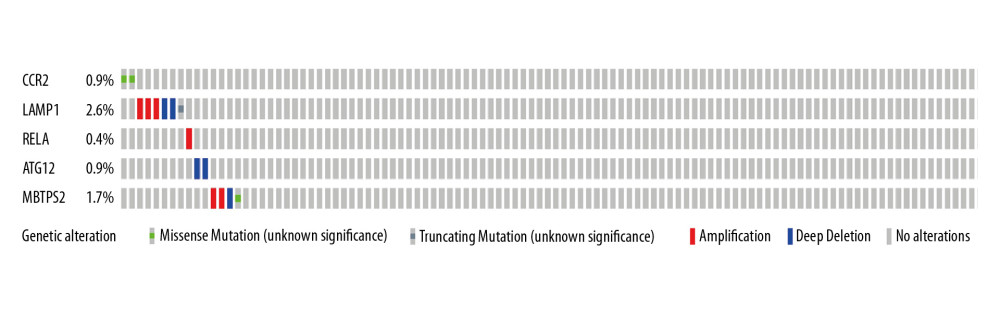 Figure 7. The mutational landscape for the 5 APRGs in LUAD patients: mutational types: amplification (red) and deletion (blue) no mutations (grey). APRGs – autophagy-pathway-related genes; LUAD – lung adenocarcinoma.
Figure 7. The mutational landscape for the 5 APRGs in LUAD patients: mutational types: amplification (red) and deletion (blue) no mutations (grey). APRGs – autophagy-pathway-related genes; LUAD – lung adenocarcinoma. References
1. Shedden K, Taylor JM, Enkemann SA, Gene expression-based survival prediction in lung adenocarcinoma: A multi-site, blinded validation study: Nat Med, 2008; 14; 822-27
2. Warth A, Muley T, Meister M, The Novel Histologic International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society Classification System of Lung Adenocarcinoma is a stage-independent predictor of survival: J Clin Oncol, 2012; 30; 1438-46
3. Arriagada R, Bergman B, Dunant A, International, cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer: N Engl J Med, 2004; 350; 351-60
4. Patel TS, Shah MG, Gandhi JS, Accuracy of cytology in sub typing non small cell lung carcinomas: Diagn Cytopathol, 2017; 45(7); 598-603
5. Molina JR, Yang P, Cassivi SD, Non-small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship: Mayo Clin Proc, 2008; 83; 584-94
6. Mehrpour M, Esclatine A, Beau I, Autophagy in health and disease. Regulation and significance of autophagy: An overview: Am J Physiol-Cellph, 2010; 298; C776-85
7. Rosenfeldt MT, Ryan KM, The multiple roles of autophagy in cancer: Carcinogenesis, 2011; 32; 955-63
8. Zhang G, Wang Z, Chen W, Dual effects of gossypol on human hepatocellular carcinoma via endoplasmic reticulum stress and autophagy: Int J Biochem Cell Biol, 2019; 113; 48-57
9. DiPrima M, Wang D, Tröster A, Identification of Eph receptor signaling as a regulator of autophagy and a therapeutic target in colorectal carcinoma: Mol Oncol, 2019; 13(11); 2441-59
10. Gil J, Ramsey D, Szmida E, The BAX gene as a candidate for negative autophagy-related genes regulator on mRNA levels in colorectal cancer: Med Oncol, 2017; 34; 16
11. Guo JY, Teng X, Laddha SV, Autophagy provides metabolic substrates to maintain energy charge and nucleotide pools in Ras-driven lung cancer cells: Genes Dev, 2016; 15; 1704-17
12. Siegel RL, Miller KD, Jemal A, Cancer statistics, 2019: Cancer J Clin, 2019; 1; 7-34
13. Ma Y, Galluzzi L, Zitvogel L, Autophagy and cellular immune responses: Immunity, 2013; 2; 211-27
14. Tini P, Belmonte G, Toscano M, Combined epidermal growth factor receptor and Beclin1 autophagic protein expression analysis identifies different clinical presentations, responses to chemo-and radiotherapy, and prognosis in glioblastoma: Biomed Res Int, 2015; 2015 208076
15. Kaverina N, Borovjagin AV, Kadagidze Z, Astrocytes promote progression of breast cancer metastases to the brain via a KISS1-mediated autophagy: Autophagy, 2017; 13; 1905-23
16. Chen D, Song Y, Zhang F, Genome-wide analysis of lung adenocarcinoma identifies novel prognostic factors and a prognostic score: Front Genet, 2019; 10; 493
17. Tsao M, Marguet S, Le Teuff G, Subtype classification of lung adenocarcinoma predicts benefit from adjuvant chemotherapy in patients undergoing complete resection: J Clin Oncol, 2015; 33; 3439-46
18. Yao M, Fang W, Smart C, CCR2 chemokine receptors enhance growth and cell-cycle progression of breast cancer cells through SRC and PKC activation: Mol Cancer Res, 2019; 17; 604-17
19. Kolattukudy PE, Niu J, Inflammation, endoplasmic reticulum stress, autophagy, and the monocyte chemoattractant protein-1/CCR2 pathway: Circ Res, 2012; 110; 174-89
20. Chen H, Li L, Hu J, UBL4A inhibits autophagy-mediated proliferation and metastasis of pancreatic ductal adenocarcinoma via targeting LAMP1: J Exp Clin Canc Res, 2019; 38; 297
21. Haller M, Hock AK, Giampazolias E, Ubiquitination and proteasomal degradation of ATG12 regulates its proapoptotic activity: Autophagy, 2015; 10(12); 2269-78
22. Sun WL, Wang L, Luo J, Ambra1 modulates the sensitivity of breast cancer cells to epirubicin by regulating autophagy via ATG12: Cancer Sci, 2018; 10; 3129-38
23. DiDonato JA, Mercurio F, Karin M, NF-κB and the link between inflammation and cancer: Immunol Rev, 2012; 246(1); 379-400
24. Lomert E, Turoverova L, Kriger D, Co-expression of RelA/p65 and ACTN4 induces apoptosis in non-small lung carcinoma cells: Cell Cycle, 2018; 5; 616-26
25. Lindert U, Cabral WA, Ausavarat S, MBTPS2 mutations cause defective regulated intramembrane proteolysis in X-linked osteogenesis imperfecta: Nat Commun, 2016; 7; 11920
26. Liu Y, Xu D, Wang L, MBTPS2 exacerbates albuminuria in streptozotocin-induced type I diabetic nephropathy by promoting endoplasmic reticulum stress-mediated renal damage: Arch Physiol Biochem, 2020 [Online ahead of print]
27. Yang Z, Xu Z, Xing H, Novel MBTPS2 mutation causes a mild phenotype of ichthyosis follicularis with atrichia and photophobia syndrome in a Chinese pedigree: J Dermatol, 2019; 46(4); e126-28
Figures
 Figure 1. The flowchart of identification of LUAD survival-related autophagy prognostic signature. Abbreviations: LUAD, lung adenocarcinoma.
Figure 1. The flowchart of identification of LUAD survival-related autophagy prognostic signature. Abbreviations: LUAD, lung adenocarcinoma. Figure 2. Differentially expressed APRGs between lung carcinoma and normal tissues. (A) The volcano plot of 5 differentially expressed APRGs. The red dots represent the level of high expression and the green dots represent the level of low expression. (B) Heatmap of 5 differently expressed APRGs. The depth of red represents the level of high expression, and the depth of green represents the level of low expression. (C) The boxplot of 5 differentially expressed APRGs. Abbreviations: APRGs, autophagy-pathway-related genes.
Figure 2. Differentially expressed APRGs between lung carcinoma and normal tissues. (A) The volcano plot of 5 differentially expressed APRGs. The red dots represent the level of high expression and the green dots represent the level of low expression. (B) Heatmap of 5 differently expressed APRGs. The depth of red represents the level of high expression, and the depth of green represents the level of low expression. (C) The boxplot of 5 differentially expressed APRGs. Abbreviations: APRGs, autophagy-pathway-related genes. Figure 3. The heat plot (A) and bubble plot (B) of enriched Gene Ontology (GO) terms. The change in color from blue to red represents the increase in the adjusted P-value, and the length of the bar indicates the number of gene enrichment terms.
Figure 3. The heat plot (A) and bubble plot (B) of enriched Gene Ontology (GO) terms. The change in color from blue to red represents the increase in the adjusted P-value, and the length of the bar indicates the number of gene enrichment terms. Figure 4. Prognostic index of 5 APRGs. (A) Lasso regression of 5 APRGs using the OS model. (B) Lasso co-efficient profiles of 5 APRGs by optimal lambda. Kaplan-Meier plot represents that patients in the high-risk group (red line) had significantly shorter overall survival time than those in the low-risk group (blue line). (C) Left: training group (D) Right: testing group. Time-dependent ROC curve analysis shows AUC values for OS in LUAD patients. (E) Left: training group. (F) Right: testing group. APRGs – autophagy-pathway-related genes; ROC – receiver-operator characteristic; AUC – area under the curve; OS – overall survival. LUAD – lung adenocarcinoma.
Figure 4. Prognostic index of 5 APRGs. (A) Lasso regression of 5 APRGs using the OS model. (B) Lasso co-efficient profiles of 5 APRGs by optimal lambda. Kaplan-Meier plot represents that patients in the high-risk group (red line) had significantly shorter overall survival time than those in the low-risk group (blue line). (C) Left: training group (D) Right: testing group. Time-dependent ROC curve analysis shows AUC values for OS in LUAD patients. (E) Left: training group. (F) Right: testing group. APRGs – autophagy-pathway-related genes; ROC – receiver-operator characteristic; AUC – area under the curve; OS – overall survival. LUAD – lung adenocarcinoma. Figure 5. Prognosis of high-risk and low-risk LUAD patients. Risk score distribution of low-risk (green) and high-risk (red) in LUAD patients in (A) training group and (B) testing group. Scatter plot of survival status of LUAD patients in (C) training group and (D) testing group. Red dots (dead); green dots (alive). Expression of risk genes in the high-risk (blue) and low-risk (pink) of the OS model in (E) training group and (F) testing group. LUAD – lung adenocarcinoma; OS – overall survival.
Figure 5. Prognosis of high-risk and low-risk LUAD patients. Risk score distribution of low-risk (green) and high-risk (red) in LUAD patients in (A) training group and (B) testing group. Scatter plot of survival status of LUAD patients in (C) training group and (D) testing group. Red dots (dead); green dots (alive). Expression of risk genes in the high-risk (blue) and low-risk (pink) of the OS model in (E) training group and (F) testing group. LUAD – lung adenocarcinoma; OS – overall survival. Figure 6. (A) Univariate and (B) multivariate analyses of OS in LUAD patients. OS – overall survival; LUAD – lung adenocarcinoma.
Figure 6. (A) Univariate and (B) multivariate analyses of OS in LUAD patients. OS – overall survival; LUAD – lung adenocarcinoma. Figure 7. The mutational landscape for the 5 APRGs in LUAD patients: mutational types: amplification (red) and deletion (blue) no mutations (grey). APRGs – autophagy-pathway-related genes; LUAD – lung adenocarcinoma.
Figure 7. The mutational landscape for the 5 APRGs in LUAD patients: mutational types: amplification (red) and deletion (blue) no mutations (grey). APRGs – autophagy-pathway-related genes; LUAD – lung adenocarcinoma. Tables
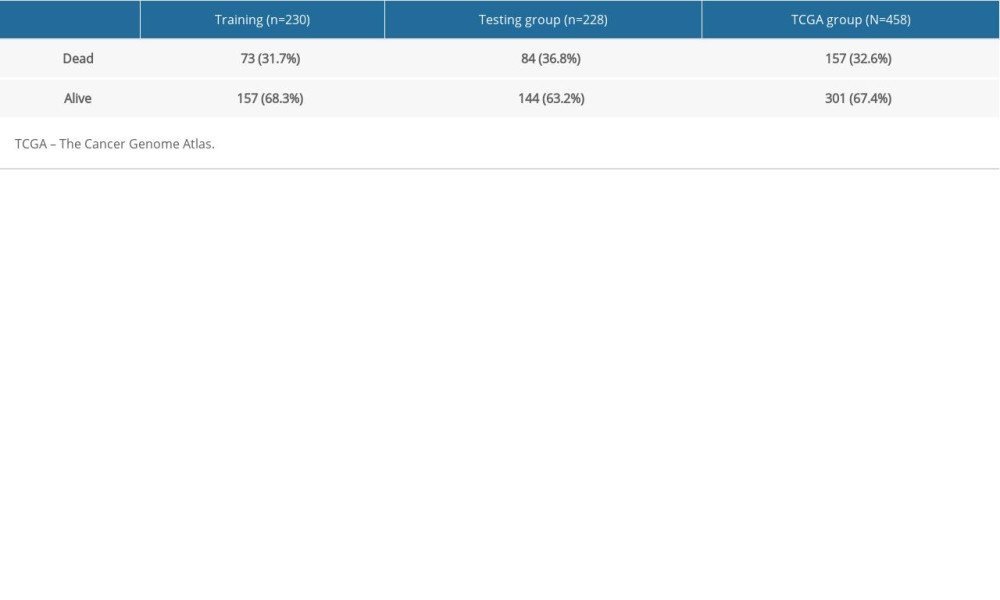 Table 1. Survival status of lung adenocarcinoma patients from different groups.
Table 1. Survival status of lung adenocarcinoma patients from different groups.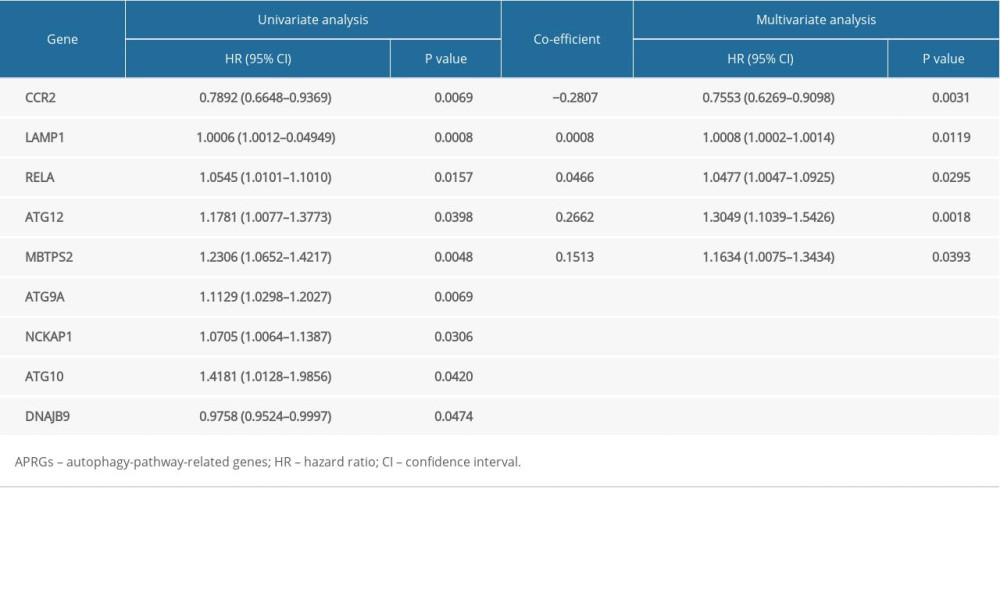 Table 2. Characteristics of risk APRGs in the prognostic model.
Table 2. Characteristics of risk APRGs in the prognostic model.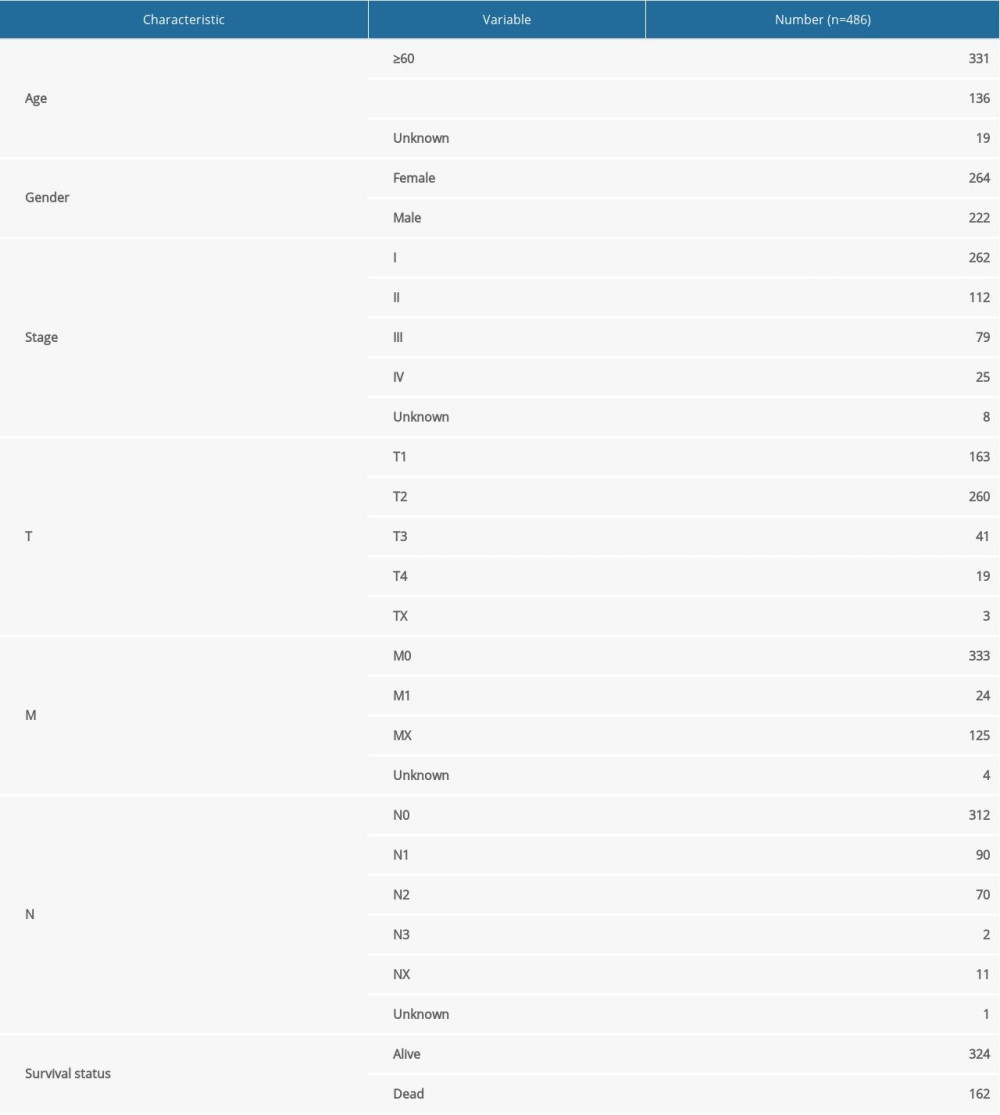 Table 3. Clinical variables of patients with lung adenocarcinoma patients.
Table 3. Clinical variables of patients with lung adenocarcinoma patients. Table 1. Survival status of lung adenocarcinoma patients from different groups.
Table 1. Survival status of lung adenocarcinoma patients from different groups. Table 2. Characteristics of risk APRGs in the prognostic model.
Table 2. Characteristics of risk APRGs in the prognostic model. Table 3. Clinical variables of patients with lung adenocarcinoma patients.
Table 3. Clinical variables of patients with lung adenocarcinoma patients. In Press
21 Mar 2024 : Meta-Analysis
Economic Evaluation of COVID-19 Screening Tests and Surveillance Strategies in Low-Income, Middle-Income, a...Med Sci Monit In Press; DOI: 10.12659/MSM.943863
10 Apr 2024 : Clinical Research
Predicting Acute Cardiovascular Complications in COVID-19: Insights from a Specialized Cardiac Referral Dep...Med Sci Monit In Press; DOI: 10.12659/MSM.942612
06 Mar 2024 : Clinical Research
Enhanced Surgical Outcomes of Popliteal Cyst Excision: A Retrospective Study Comparing Arthroscopic Debride...Med Sci Monit In Press; DOI: 10.12659/MSM.941102
06 Mar 2024 : Clinical Research
Prevalence and Variation of Medical Comorbidities in Oral Surgery Patients: A Retrospective Study at Jazan ...Med Sci Monit In Press; DOI: 10.12659/MSM.943884
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952








