18 February 2021: Database Analysis
Prognostic Significance of the Hsp70 Gene Family in Colorectal Cancer
Wenjun Jiang1AC, Xiaohang Pan2BF, Haifan Yan1EF, Guoping Wang1AD*DOI: 10.12659/MSM.928352
Med Sci Monit 2021; 27:e928352
Abstract
BACKGROUND: Colorectal cancer (CRC) is a deadly form of cancer worldwide. Heat shock protein 70 (Hsp70) belongs to the family of human HSPs and plays an essential role in multiple cellular developments and in responding to environmental changes. However, studies on the relationship between CRC and the Hsp70 family are rare.
MATERIAL AND METHODS: Data pertaining to 438 patients with CRC was downloaded from The Cancer Genome Atlas database. To investigate the prognostic significance of the Hsp70 genes, survival and joint-effect analyses were conducted. The correlation between prognosis-related Hsp70 genes and clinical factors in CRC was analyzed using a nomogram. Gene set enrichment analysis (GSEA) was performed to explore the complex enrichment pathway in CRC with the prognosis-related Hsp70 genes.
RESULTS: According to multivariate Cox regression survival analysis, low expression levels of HSPA1A, HSPA1B, and HSPA1L were correlated with improved overall survival (OS). According to the joint-effects survival analysis, the joint low expression levels of HSPA1A, HSPA1B, and HSPA1L were related to improved OS. The 1-, 3-, 5-, and 10-year survival rates of patients with CRC were predicted by constructing a nomogram model based on HSPA1A, HSPA1B, HSPA1L, and tumor stage. The GSEA results indicated the biological roles of HSPA1A, HSPA1B, and HSPA1L in CRC.
CONCLUSIONS: Low expression levels of HSPA1A, HSPA1B, and HSPA1L were strongly correlated with improved prognosis in CRC and might serve as latent prognostic biomarkers in CRC.
Keywords: Colorectal Neoplasms, Hereditary Nonpolyposis, HSP70 Heat-Shock Proteins, Case-Control Studies, Colorectal Neoplasms, Databases, Genetic, Genetic Predisposition to Disease, Genotype, nomograms, Polymorphism, Single Nucleotide
Background
Colorectal cancer (CRC) causes many fatalities worldwide. However, timely diagnosis and therapy can slow the progression of CRC [1]. It has been found that during advanced stages of CRC, when metastasis has commenced, carcinoembryonic antigen (CEA) and carbohydrate antigen 199 (CA199) are elevated, and these antigen levels are being utilized in clinical practice with limited effectiveness [2].
Molecular chaperones aid in the dissolution of misfolded proteins. Consequently, they fulfill a crucial physiological role [3]. Heat shock protein 70 (
Studies have demonstrated that
Material and Methods
DATA PREPARATION:
The TCGA dataset is a substantial network database for researchers (https://cancergenome.nih.gov/), which stores information on different genomes of primary tumors and matched normal tissues [11]. In this study, we analyzed data from 438 patients with CRC, which included Hsp70 gene family expression and clinical data. Scatter plots were generated for the Hsp70 gene family in CRC and matched normal tissues.
INTERACTION AND FUNCTION ANALYSIS OF THE HSP70 GENE FAMILY:
The Pearson correlation coefficient was used for the correlation analysis of Hsp70 genes. The coexpression correlation of Hsp70 genes was performed in GeneMANIA (www.genemania.org) [12]. The functional bioinformatics analysis of Hsp70 genes was conducted using the online tool DAVID (david.ncifcrf.gov/tools.jsp) [13].
SURVIVAL AND JOINT-EFFECT ANALYSIS OF THE HSP70 GENE FAMILY:
Univariate and multivariate Cox proportional hazard ratios (HRs) were used to determine the effects of all
NOMOGRAM:
A nomogram was formulated for the prognosis-related Hsp70 genes and clinical factors in CRC. The 1-year, 3-year, 5-year, and 10-year survival rates in CRC patients were predicted using the nomogram [14].
GENE SET ENRICHMENT ANALYSIS:
Gene set enrichment analysis (GSEA) v.3.0 (http://software.broadinstitute.org/gsea/msigdb/index.jsp) was used to analyze the enrichment pathway in CRC with the prognosis-related Hsp70 genes [15]. The Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) datasets were used for analysis. Statistical significance was indicated by P<0.05 and a false discovery rate <0.25.
STATISTICAL ANALYSIS:
SPSS version 25.0 (IBM, Chicago, IL, USA) was used for statistical analysis. The calculation of survival analysis was carried out with Cox proportional hazards regression and Kaplan-Meier analyses, which yielded log-rank
Results
CLINICAL CHARACTERISTICS:
The clinical data from 438 patients with CRC was used in the analysis. Table 1 lists the correlations between clinical characteristics and OS in the patients with CRC [16]. The results showed that TNM stage was related with OS. Figure 1 illustrates each of the Hsp70 family gene levels in samples of CRC and normal colon tissue. HSPA1A, HSPA1B, HSPA1L, HSPA2, HSPA4, HSPA4L, HSPA5, HSPA8, HSPA9, HSPA12B, HSPA14, HSPH1, and HYOU1 gene levels were statistically significant.
INTERACTION AND FUNCTION ANALYSIS OF THE HSP70 GENE FAMILY:
The Pearson correlation coefficient was used to analyze the correlation of Hsp70 genes (Figure 2). Figure 3 illustrates the gene-gene and protein–protein interaction network of the Hsp70 gene family. Figure 4 illustrates the GO pathway functional analysis and KEGG pathway functional analysis.
SURVIVAL AND JOINT EFFECT ANALYSIS OF THE HSP70 GENE FAMILY:
Table 2 summarizes the univariate and multivariate survival analyses of the Hsp70 genes. Low expression levels of HSPA1A, HSPA1B, and HSPA1L were associated with improved OS in univariate survival analysis. Meanwhile, the elevated expression level of HSPA9 was related to improved OS (Figure 5). Moreover, multivariate survival analysis demonstrated that lower expression levels of HSPA1A, HSPA1B, and HSPA1L were significantly associated with improved OS.
Based on the multivariate survival analysis of HSPA1A, HSPA1B, and HSPA1L, a joint-effects framework was performed with different groups (Table 3). As illustrated in Figure 6, low expression levels of HSPA1A, HSPA1B, and HSPA1L in Groups 1, 4, 7, and 10 were significantly correlated with improved OS.
NOMOGRAM:
The nomogram was utilized for investigating the association between HSPA1A, HSPA1B, HSPA1L, and tumor stage in CRC. The points of each variable could be calculated. Figure 7 shows the prediction of the 1-, 3-, 5-, and 10-year survival rates.
GSEA ANALYSIS:
To investigate the enrichment pathway with HSPA1A, HSPA1B, and HSPA1L, GSEA analysis was conducted. As illustrated in Figure 8, according to the GSEA, the low expression of HSPA1A was positively correlated with the cell cycle, DNA replication, RNA degradation, and P53 pathway. As illustrated in Figure 9, the GSEA indicated that the high expression of HSPA1B was positively correlated with the spliceosome, heat shock protein binding, RNA polymerase II promoter transcription elongation, DNA-templated transcription elongation, chaperone-mediated protein folding, and positive regulation of gene-expression epigenetics. The GSEA also indicated that a low expression of HSPA1L was positively correlated with the cell cycle, DNA replication, DNA helicase activity, and P53 pathway (Figure 10).
Discussion
The TCGA database was utilized to illustrate the importance of
Since the
It has been demonstrated that
Similar to
This study has a number of limitations. First, the public databases lack detailed clinical information. Second, the patient data were obtained from a single source. To generalize the results, it will be necessary to validate the conclusions through the analysis of independent data in future studies. Finally, since this study is mainly a bioinformatics study using data from a public database, it lacks empirical conclusiveness. The anticancer properties of
Conclusions
Through comprehensive analysis, we identified the potential molecular mechanisms of
Figures
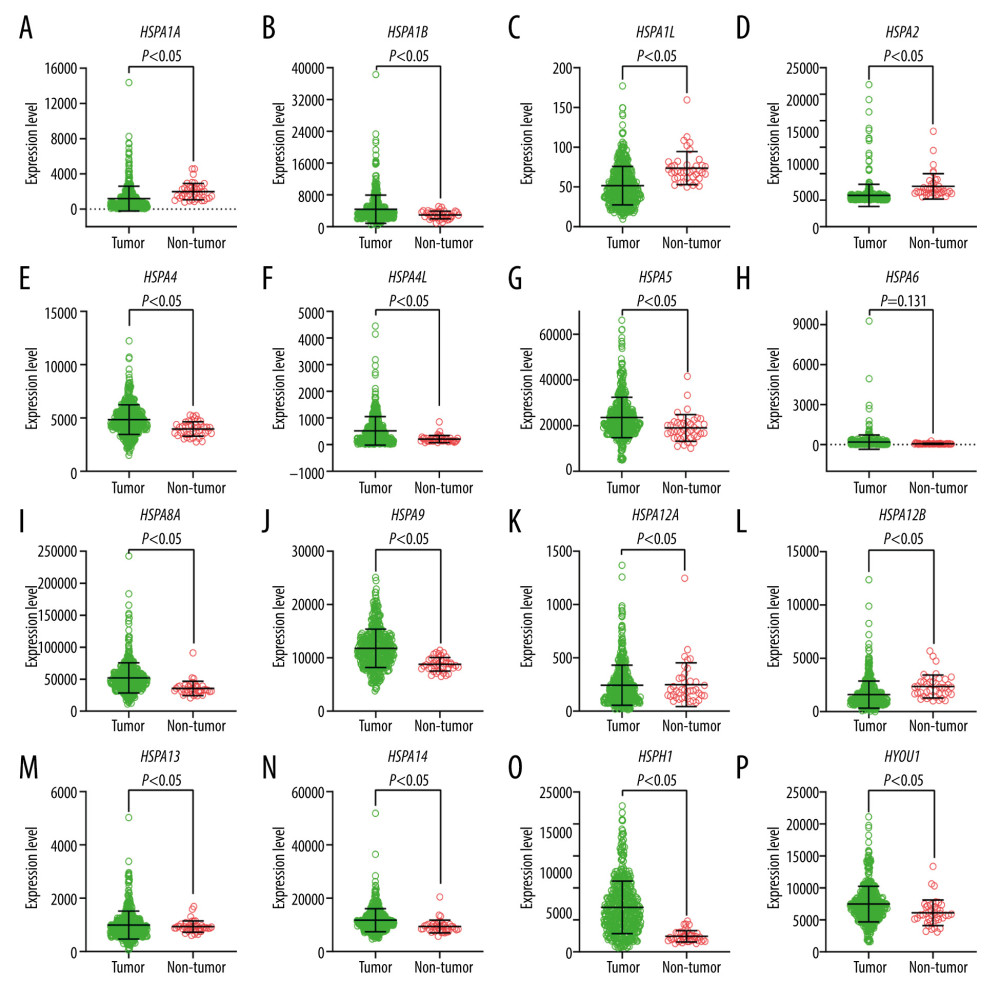 Figure 1. Expression levels of Hsp70 genes in colorectal cancer and normal colon tissue. (A) HSPA1A; (B) HSPA1B; (C) HSPA1L; (D) HSPA2; (E) HSPA4; (F) HSPA4L; (G) HSPA5; (H) HSPA6; (I) HSPA8; (J) HSPA9; (K) HSPA12A; (L) HSPA12B; (M) HSPA13; (N) HSPA14; (O) HSPH1; (P) HYOU1.
Figure 1. Expression levels of Hsp70 genes in colorectal cancer and normal colon tissue. (A) HSPA1A; (B) HSPA1B; (C) HSPA1L; (D) HSPA2; (E) HSPA4; (F) HSPA4L; (G) HSPA5; (H) HSPA6; (I) HSPA8; (J) HSPA9; (K) HSPA12A; (L) HSPA12B; (M) HSPA13; (N) HSPA14; (O) HSPH1; (P) HYOU1. 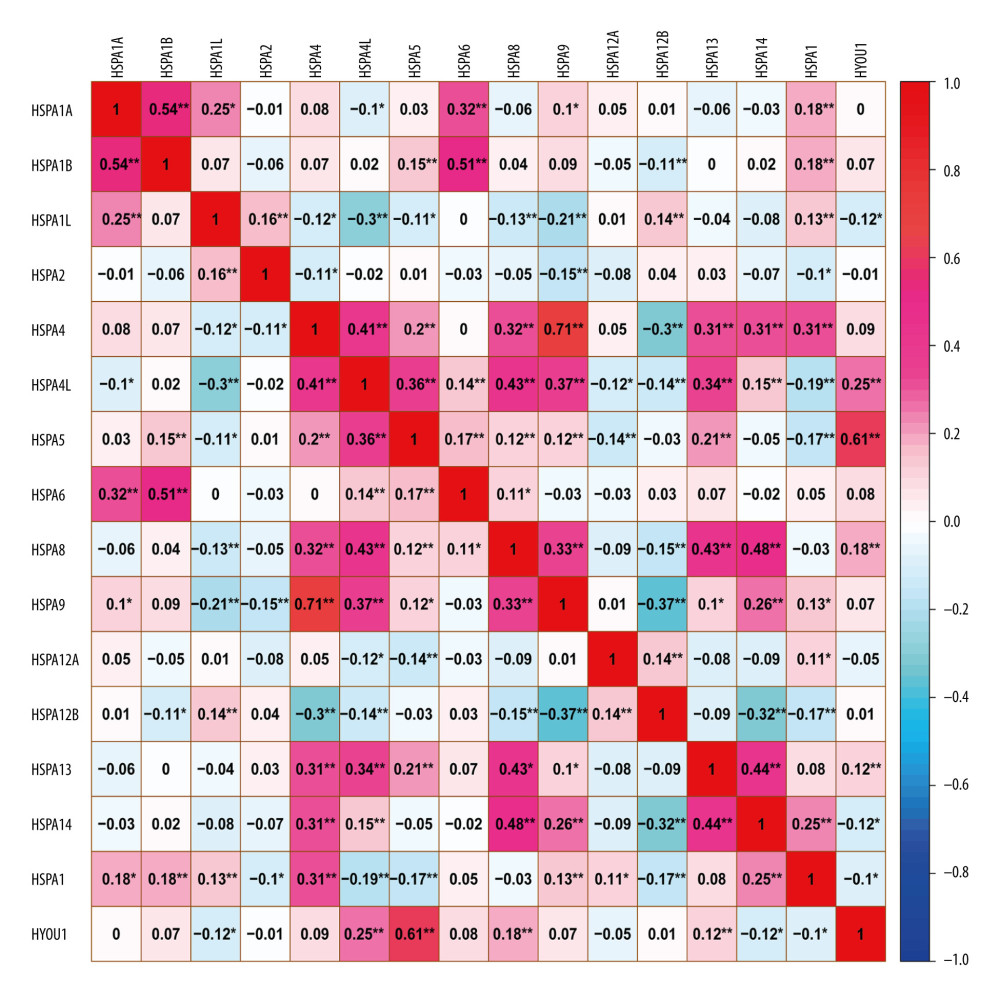 Figure 2. Pearson’s correlation analysis for HSPA1A, HSPA1B, HSPA1L, HSPA2, HSPA4, HSPA4L, HSPA5, HSPA6, HSPA8, HSPA9, HSPA12A, HSPA12B, HSPA13, HSPA14, HSPH1, and HYOU1.
Figure 2. Pearson’s correlation analysis for HSPA1A, HSPA1B, HSPA1L, HSPA2, HSPA4, HSPA4L, HSPA5, HSPA6, HSPA8, HSPA9, HSPA12A, HSPA12B, HSPA13, HSPA14, HSPH1, and HYOU1. 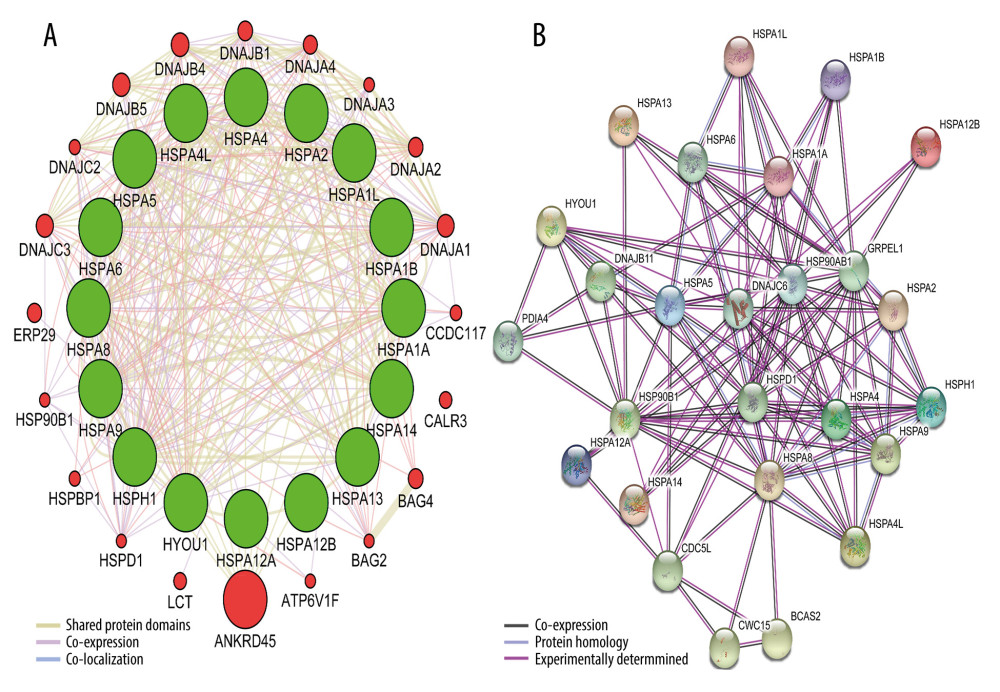 Figure 3. Gene-gene and protein–protein interaction network for Hsp70 gene family. (A) Gene-gene interaction network; (B) Protein–protein interaction network.
Figure 3. Gene-gene and protein–protein interaction network for Hsp70 gene family. (A) Gene-gene interaction network; (B) Protein–protein interaction network. 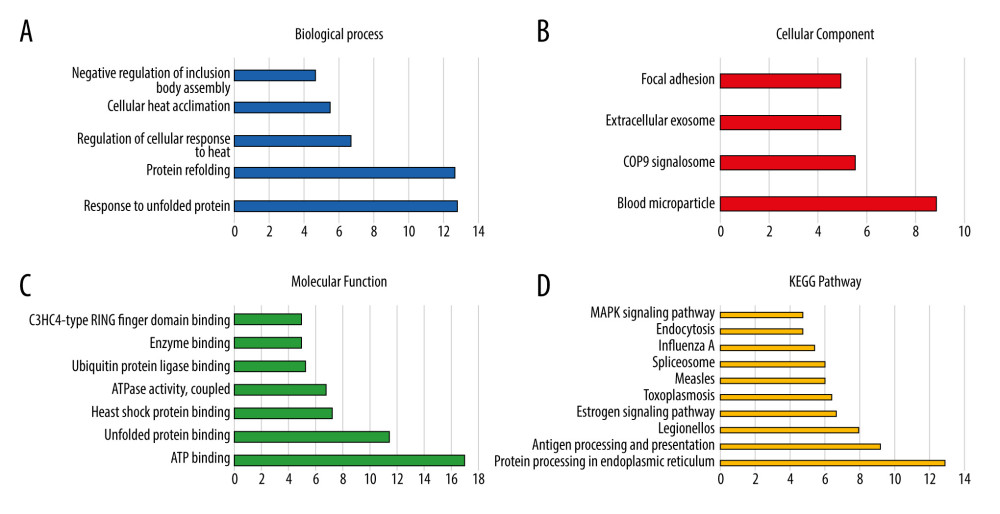 Figure 4. GO and KEGG pathway analysis of Hsp70 gene family carried out by the online tool DAVID. (A) Biological process; (B) cellular component; (C) molecular function; (D) KEGG pathway.
Figure 4. GO and KEGG pathway analysis of Hsp70 gene family carried out by the online tool DAVID. (A) Biological process; (B) cellular component; (C) molecular function; (D) KEGG pathway. 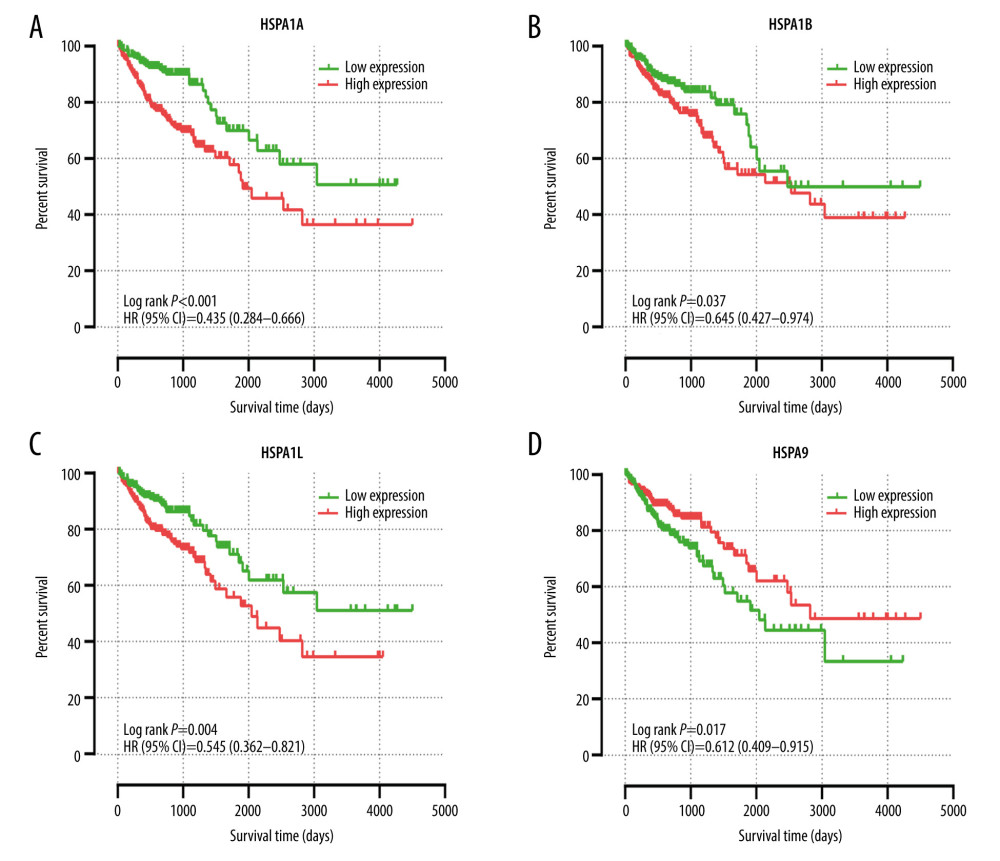 Figure 5. The univariate survival analysis of HSPA1A, HSPA1B, HSPA1L, and HSPA9 (P<0.05). Kaplan-Meier survival curves concerning (A) HSPA1A; (B) HSPA1B; (C) HSPA1L; (D) HSPA9 expression.
Figure 5. The univariate survival analysis of HSPA1A, HSPA1B, HSPA1L, and HSPA9 (P<0.05). Kaplan-Meier survival curves concerning (A) HSPA1A; (B) HSPA1B; (C) HSPA1L; (D) HSPA9 expression. 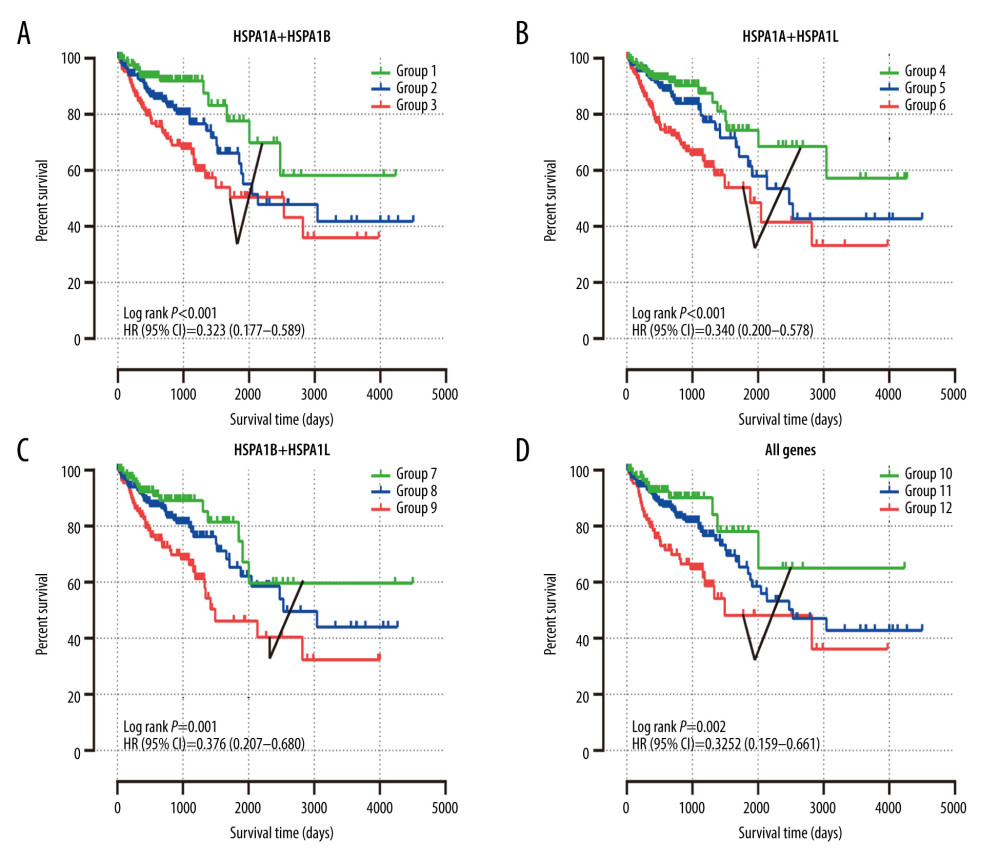 Figure 6. The joint-effects analysis of the influence of combined HSPA1A, HSPA1B, and HSPA1L. Kaplan-Meier survival curves concerning (A) HSPA1A+HSPA1B; (B) HSPA1A+HSPA1L; (C) HSPA1B+HSPA1L; (D) HSPA1A+HSPA1B+HSPA1L.
Figure 6. The joint-effects analysis of the influence of combined HSPA1A, HSPA1B, and HSPA1L. Kaplan-Meier survival curves concerning (A) HSPA1A+HSPA1B; (B) HSPA1A+HSPA1L; (C) HSPA1B+HSPA1L; (D) HSPA1A+HSPA1B+HSPA1L. 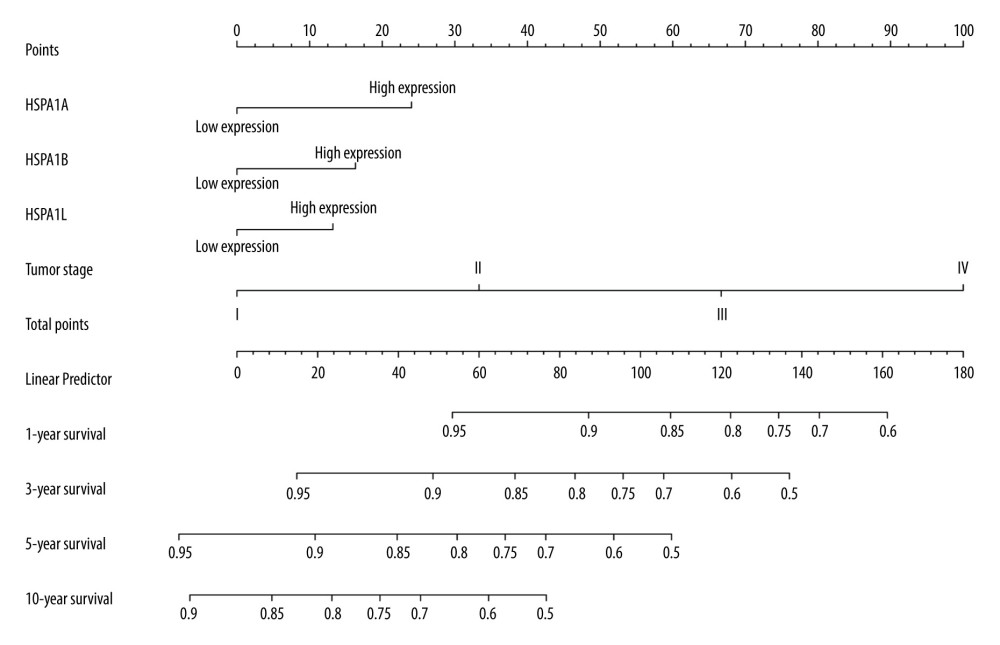 Figure 7. A nomogram model was performed to analyze the prognosis correlation of HSPA1A, HSPA1B, HSPA1L and tumor stage in CRC. The points of each variable were calculated at the top of the nomogram. A vertical line down to the 1-, 3-, 5-, and 10-year survival lines allowed for the determination of survival probabilities.
Figure 7. A nomogram model was performed to analyze the prognosis correlation of HSPA1A, HSPA1B, HSPA1L and tumor stage in CRC. The points of each variable were calculated at the top of the nomogram. A vertical line down to the 1-, 3-, 5-, and 10-year survival lines allowed for the determination of survival probabilities. 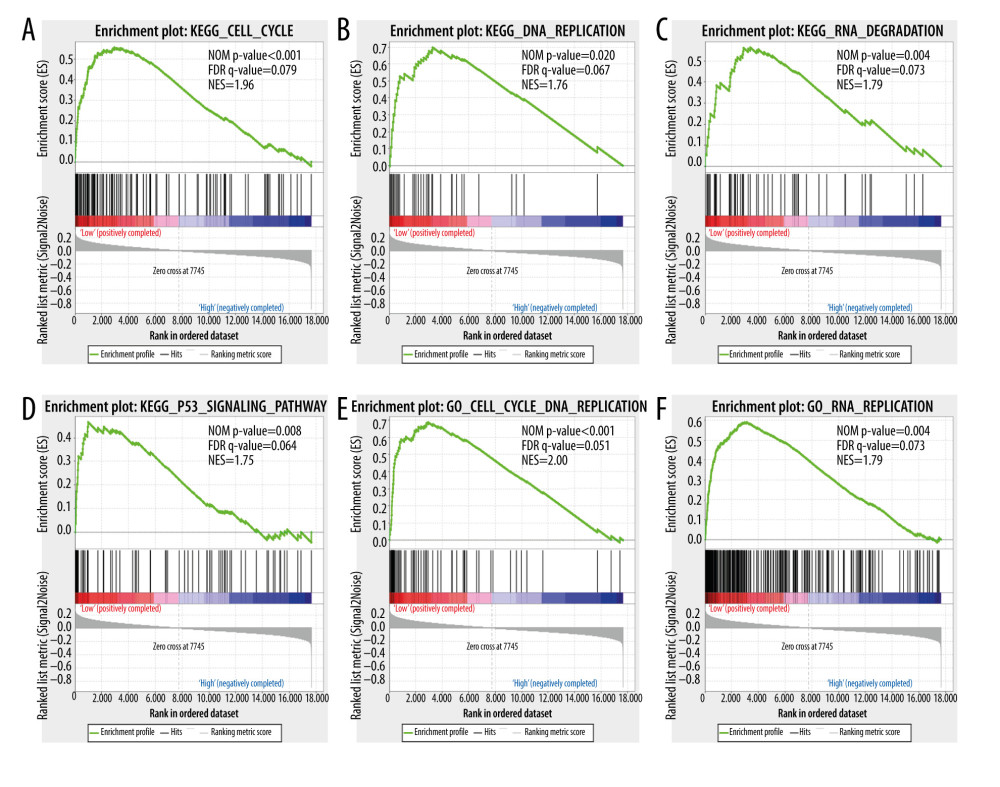 Figure 8. Gene set enrichment analysis shows the enrichment analysis of HSPA1A. (A–F) Statistical significance was implied by NOM P<0.05 and FDR<0.25. NOM – normalized; FDR – false discovery rate; NES – normalized enrichment score.
Figure 8. Gene set enrichment analysis shows the enrichment analysis of HSPA1A. (A–F) Statistical significance was implied by NOM P<0.05 and FDR<0.25. NOM – normalized; FDR – false discovery rate; NES – normalized enrichment score. 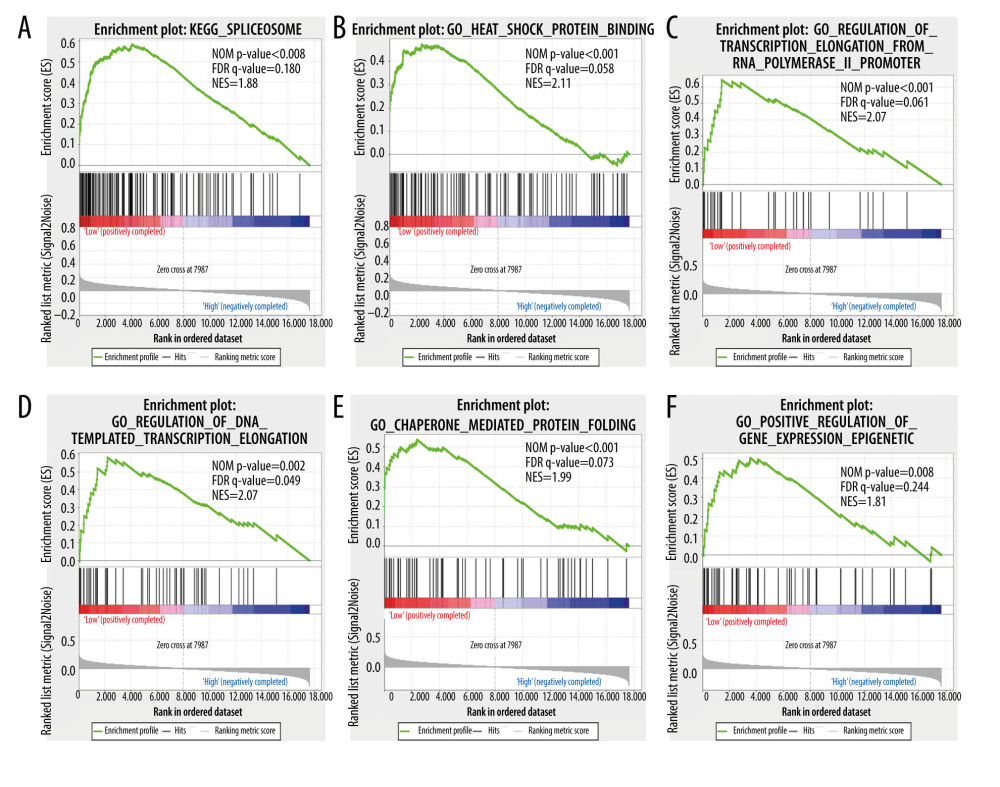 Figure 9. Gene set enrichment analysis shows the enrichment analysis of HSPA1B. (A–F) Statistical significance was implied by NOM P<0.05 and FDR<0.25. NOM – normalized; FDR – false discovery rate; NES – normalized enrichment score.
Figure 9. Gene set enrichment analysis shows the enrichment analysis of HSPA1B. (A–F) Statistical significance was implied by NOM P<0.05 and FDR<0.25. NOM – normalized; FDR – false discovery rate; NES – normalized enrichment score. 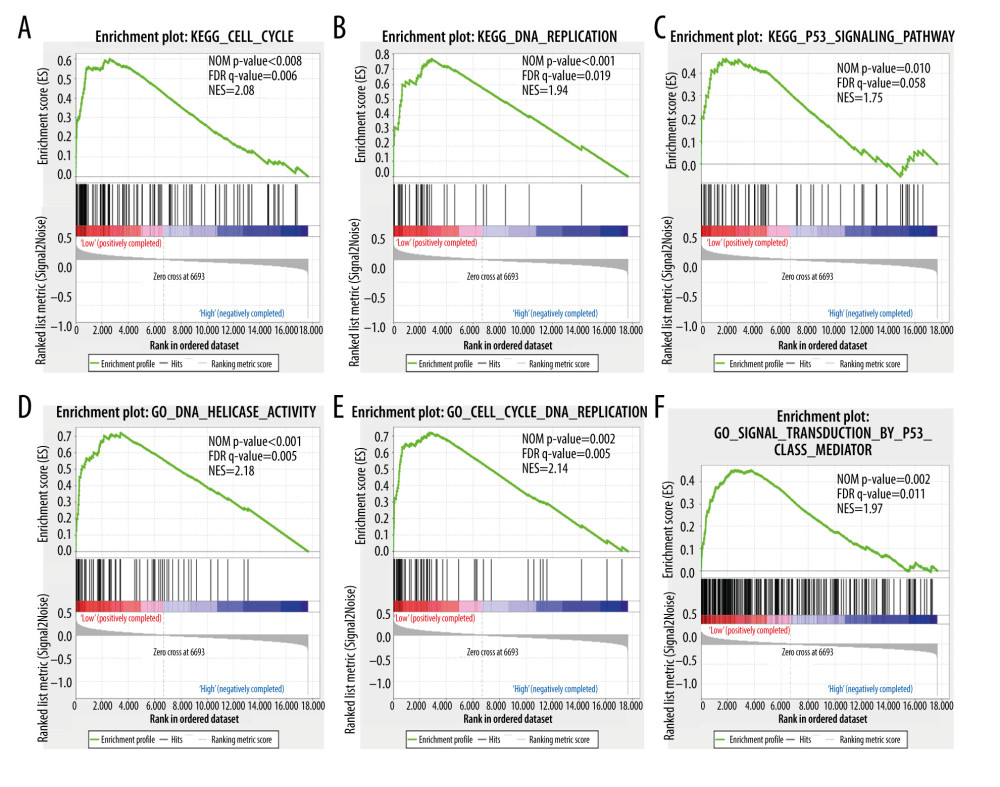 Figure 10. Gene set enrichment analysis shows the enrichment analysis of HSPA1L. (A–F) Statistical significance was implied by NOM P<0.05 and FDR<0.25. NOM – normalized; FDR – false discovery rate; NES – normalized enrichment score.
Figure 10. Gene set enrichment analysis shows the enrichment analysis of HSPA1L. (A–F) Statistical significance was implied by NOM P<0.05 and FDR<0.25. NOM – normalized; FDR – false discovery rate; NES – normalized enrichment score. References
1. Siegel RL, Miller KD, Jemal A, Cancer statistics, 2020: Cancer J Clin, 2020; 70; 7-30
2. Kannagi R, Izawa M, Koike T, Carbohydrate-mediated cell adhesion in cancer metastasis and angiogenesis: Cancer Sci, 2004; 95; 377-84
3. Brodsky JL, Chiosis G, Hsp70 molecular chaperones: Emerging roles in human disease and identification of small molecule modulators: Curr Top Med Chem, 2006; 6; 1215-25
4. Daugaard M, Rohde M, Jaattela M, The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions: FEBS Letters, 2007; 581; 3702-10
5. Nylandsted J, Brand K, Jäättelä M, Heat shock protein 70 is required for the survival of cancer cells: Ann N Y Acad Sci, 2000; 926; 122-25
6. Kumar S, Stokes J, Singh UP, Targeting Hsp70: A possible therapy for cancer: Cancer Lett, 2016; 374; 156-66
7. Radons J, The human HSP70 family of chaperones: Where do we stand?: Cell Stress Chaperon, 2016; 21; 379-404
8. Brocchieri L, Conway de Macario E, Macario AJ, Hsp70 genes in the human genome: Conservation and differentiation patterns predict a wide array of overlapping and specialized functions: BMC Evol Biol, 2008; 8; 19
9. Bauer K, Nitsche U, Slotta-Huspenina J, High HSP27 and HSP70 expression levels are independent adverse prognostic factors in primary resected colon cancer: Cell Oncol (Dordrecht), 2012; 35; 197-205
10. Soleimani A, Zahiri E, Ehtiati S, Therapeutic potency of heat-shock protein-70 in the pathogenesis of colorectal cancer: Current status and perspectives: Biochem Cell Biol, 2019; 97; 85-90
11. Weinstein JN, Collisson EA, Mills GB, The Cancer Genome Atlas Pan-Cancer analysis project: Nat Genet, 2013; 45; 1113-20
12. Warde-Farley D, Donaldson SL, Comes O, The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function: Nucleic Acids Res, 2010; 38; W214-20
13. Huang DW, Sherman BT, Lempicki RA, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources: Nat Protoc, 2009; 4; 44-57
14. Balachandran VP, Gonen M, Smith JJ, DeMatteo RP, Nomograms in oncology: More than meets the eye: Lancet Oncol, 2015; 16; e173-80
15. Subramanian A, Tamayo P, Mootha VK, Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles: Proc Natl Acad Sci USA, 2005; 102; 15545-50
16. Pan X, Wang Q, Xu C, Prognostic value of chloride channel accessory mRNA expression in colon cancer: Oncol Lett, 2019; 18; 2967-76
17. Leung TK, Hall C, Rajendran M, The human heat-shock genes HSPA6 and HSPA7 are both expressed and localize to chromosome 1: Genomics, 1992; 12; 74-79
18. Kudla G, Helwak A, Lipinski L, Gene conversion and GC-content evolution in mammalian Hsp70: Mol Biol Evol, 2004; 21; 1438-44
19. Guo H, Deng Q, Wu C, Variations in HSPA1B at 6p21.3 are associated with lung cancer risk and prognosis in Chinese populations: Cancer Res, 2011; 71; 7576-86
20. Kampinga HH, Hageman J, Vos MJ, Guidelines for the nomenclature of the human heat shock proteins: Cell Stress Chaperon, 2009; 14; 105-11
21. Garrido C, Brunet M, Didelot C, Heat shock proteins 27 and 70: Anti-apoptotic proteins with tumorigenic properties: Cell Cycle (Georgetown, Tex), 2006; 5; 2592-601
22. Klink M, Nowak M, Kielbik M: Cell Stress Chaperon, 2012; 17; 661-74
23. Nadin SB, Cuello-Carrión FD, Sottile ML, Effects of hyperthermia on Hsp27 (HSPB1), Hsp72 (HSPA1A) and DNA repair proteins hMLH1 and hMSH2 in human colorectal cancer hMLH1-deficient and hMLH1-proficient cell lines: Int J Hyperthermia, 2012; 28; 191-201
24. Ban Y, Tan P, Cai J, LNCAROD is stabilized by m6A methylation and promotes cancer progression via forming a ternary complex with HSPA1A and YBX1 in head and neck squamous cell carcinoma: Mol Oncol, 2020; 14; 1282-96
25. De Andrade WP, Da Conceição Braga L, Gonçales NG, HSPA1A, HSPA1L and TRAP1 heat shock genes may be associated with prognosis in ovarian epithelial cancer: Oncol Lett, 2020; 19; 359-67
26. Jagadish N, Parashar D, Gupta N, Heat shock protein 70-2 (HSP70-2) is a novel therapeutic target for colorectal cancer and is associated with tumor growth: BMC Cancer, 2016; 16; 561
27. Jagadish N, Agarwal S, Gupta N, Heat shock protein 70-2 (HSP70-2) overexpression in breast cancer: J Exp Clin Cancer Res, 2016; 35; 150
28. Sfar S, Saad H, Mosbah F, Chouchane L, Association of HSP70-hom genetic variant with prostate cancer risk: Mol Biol Rep, 2008; 35; 459-64
29. Mosser DD, Caron AW, Bourget L, The chaperone function of hsp70 is required for protection against stress-induced apoptosis: Mol Cell Biol, 2000; 20; 7146-59
30. Nylandsted J, Rohde M, Brand K, Selective depletion of heat shock protein 70 (Hsp70) activates a tumor-specific death program that is independent of caspases and bypasses Bcl-2: Proc Natl Acad Sci USA, 2000; 97; 7871-76
31. Jäättelä M, Wissing D, Kokholm K, Hsp70 exerts its anti-apoptotic function downstream of caspase-3-like proteases: EMBO J, 1998; 17; 6124-34
32. Kalinowska M, Garncarz W, Pietrowska M, Regulation of the human apoptotic DNase/RNase endonuclease G: Involvement of Hsp70 and ATP: Apoptosis, 2005; 10; 821-30
33. Kang Y, Jung WY, Lee H, Prognostic significance of heat shock protein 70 expression in early gastric carcinoma: Korean J Pathol, 2013; 47; 219-26
34. Dimas DT, Perlepe CD, Sergentanis TN, The prognostic significance of Hsp70/Hsp90 expression in breast cancer: A systematic review and meta-analysis: Anticancer Res, 2018; 38; 1551-62
Figures
 Figure 1. Expression levels of Hsp70 genes in colorectal cancer and normal colon tissue. (A) HSPA1A; (B) HSPA1B; (C) HSPA1L; (D) HSPA2; (E) HSPA4; (F) HSPA4L; (G) HSPA5; (H) HSPA6; (I) HSPA8; (J) HSPA9; (K) HSPA12A; (L) HSPA12B; (M) HSPA13; (N) HSPA14; (O) HSPH1; (P) HYOU1.
Figure 1. Expression levels of Hsp70 genes in colorectal cancer and normal colon tissue. (A) HSPA1A; (B) HSPA1B; (C) HSPA1L; (D) HSPA2; (E) HSPA4; (F) HSPA4L; (G) HSPA5; (H) HSPA6; (I) HSPA8; (J) HSPA9; (K) HSPA12A; (L) HSPA12B; (M) HSPA13; (N) HSPA14; (O) HSPH1; (P) HYOU1. Figure 2. Pearson’s correlation analysis for HSPA1A, HSPA1B, HSPA1L, HSPA2, HSPA4, HSPA4L, HSPA5, HSPA6, HSPA8, HSPA9, HSPA12A, HSPA12B, HSPA13, HSPA14, HSPH1, and HYOU1.
Figure 2. Pearson’s correlation analysis for HSPA1A, HSPA1B, HSPA1L, HSPA2, HSPA4, HSPA4L, HSPA5, HSPA6, HSPA8, HSPA9, HSPA12A, HSPA12B, HSPA13, HSPA14, HSPH1, and HYOU1. Figure 3. Gene-gene and protein–protein interaction network for Hsp70 gene family. (A) Gene-gene interaction network; (B) Protein–protein interaction network.
Figure 3. Gene-gene and protein–protein interaction network for Hsp70 gene family. (A) Gene-gene interaction network; (B) Protein–protein interaction network. Figure 4. GO and KEGG pathway analysis of Hsp70 gene family carried out by the online tool DAVID. (A) Biological process; (B) cellular component; (C) molecular function; (D) KEGG pathway.
Figure 4. GO and KEGG pathway analysis of Hsp70 gene family carried out by the online tool DAVID. (A) Biological process; (B) cellular component; (C) molecular function; (D) KEGG pathway. Figure 5. The univariate survival analysis of HSPA1A, HSPA1B, HSPA1L, and HSPA9 (P<0.05). Kaplan-Meier survival curves concerning (A) HSPA1A; (B) HSPA1B; (C) HSPA1L; (D) HSPA9 expression.
Figure 5. The univariate survival analysis of HSPA1A, HSPA1B, HSPA1L, and HSPA9 (P<0.05). Kaplan-Meier survival curves concerning (A) HSPA1A; (B) HSPA1B; (C) HSPA1L; (D) HSPA9 expression. Figure 6. The joint-effects analysis of the influence of combined HSPA1A, HSPA1B, and HSPA1L. Kaplan-Meier survival curves concerning (A) HSPA1A+HSPA1B; (B) HSPA1A+HSPA1L; (C) HSPA1B+HSPA1L; (D) HSPA1A+HSPA1B+HSPA1L.
Figure 6. The joint-effects analysis of the influence of combined HSPA1A, HSPA1B, and HSPA1L. Kaplan-Meier survival curves concerning (A) HSPA1A+HSPA1B; (B) HSPA1A+HSPA1L; (C) HSPA1B+HSPA1L; (D) HSPA1A+HSPA1B+HSPA1L. Figure 7. A nomogram model was performed to analyze the prognosis correlation of HSPA1A, HSPA1B, HSPA1L and tumor stage in CRC. The points of each variable were calculated at the top of the nomogram. A vertical line down to the 1-, 3-, 5-, and 10-year survival lines allowed for the determination of survival probabilities.
Figure 7. A nomogram model was performed to analyze the prognosis correlation of HSPA1A, HSPA1B, HSPA1L and tumor stage in CRC. The points of each variable were calculated at the top of the nomogram. A vertical line down to the 1-, 3-, 5-, and 10-year survival lines allowed for the determination of survival probabilities. Figure 8. Gene set enrichment analysis shows the enrichment analysis of HSPA1A. (A–F) Statistical significance was implied by NOM P<0.05 and FDR<0.25. NOM – normalized; FDR – false discovery rate; NES – normalized enrichment score.
Figure 8. Gene set enrichment analysis shows the enrichment analysis of HSPA1A. (A–F) Statistical significance was implied by NOM P<0.05 and FDR<0.25. NOM – normalized; FDR – false discovery rate; NES – normalized enrichment score. Figure 9. Gene set enrichment analysis shows the enrichment analysis of HSPA1B. (A–F) Statistical significance was implied by NOM P<0.05 and FDR<0.25. NOM – normalized; FDR – false discovery rate; NES – normalized enrichment score.
Figure 9. Gene set enrichment analysis shows the enrichment analysis of HSPA1B. (A–F) Statistical significance was implied by NOM P<0.05 and FDR<0.25. NOM – normalized; FDR – false discovery rate; NES – normalized enrichment score. Figure 10. Gene set enrichment analysis shows the enrichment analysis of HSPA1L. (A–F) Statistical significance was implied by NOM P<0.05 and FDR<0.25. NOM – normalized; FDR – false discovery rate; NES – normalized enrichment score.
Figure 10. Gene set enrichment analysis shows the enrichment analysis of HSPA1L. (A–F) Statistical significance was implied by NOM P<0.05 and FDR<0.25. NOM – normalized; FDR – false discovery rate; NES – normalized enrichment score. Tables
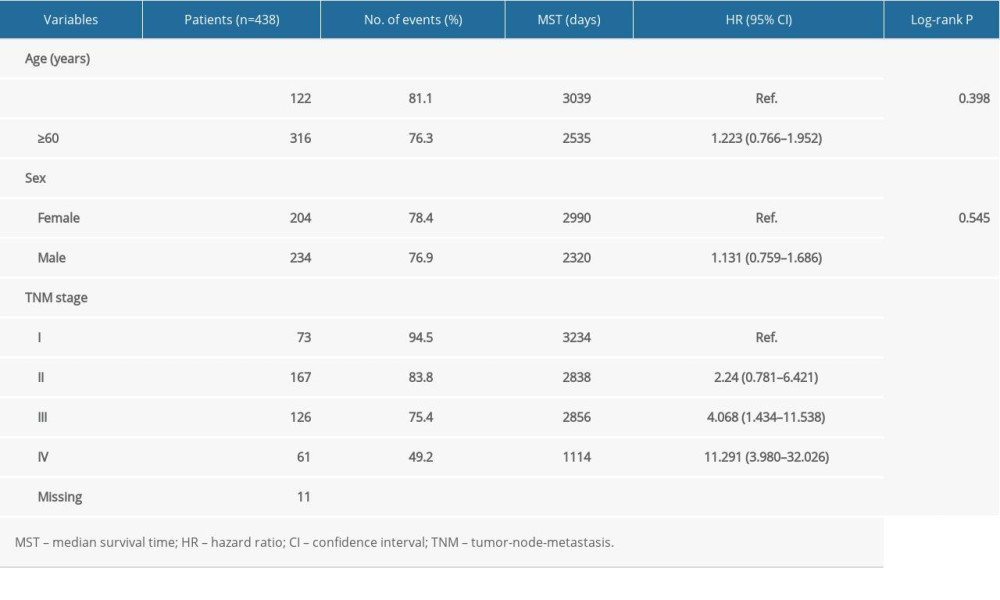 Table 1. The clinical data for 438 patients with colorectal cancer.
Table 1. The clinical data for 438 patients with colorectal cancer.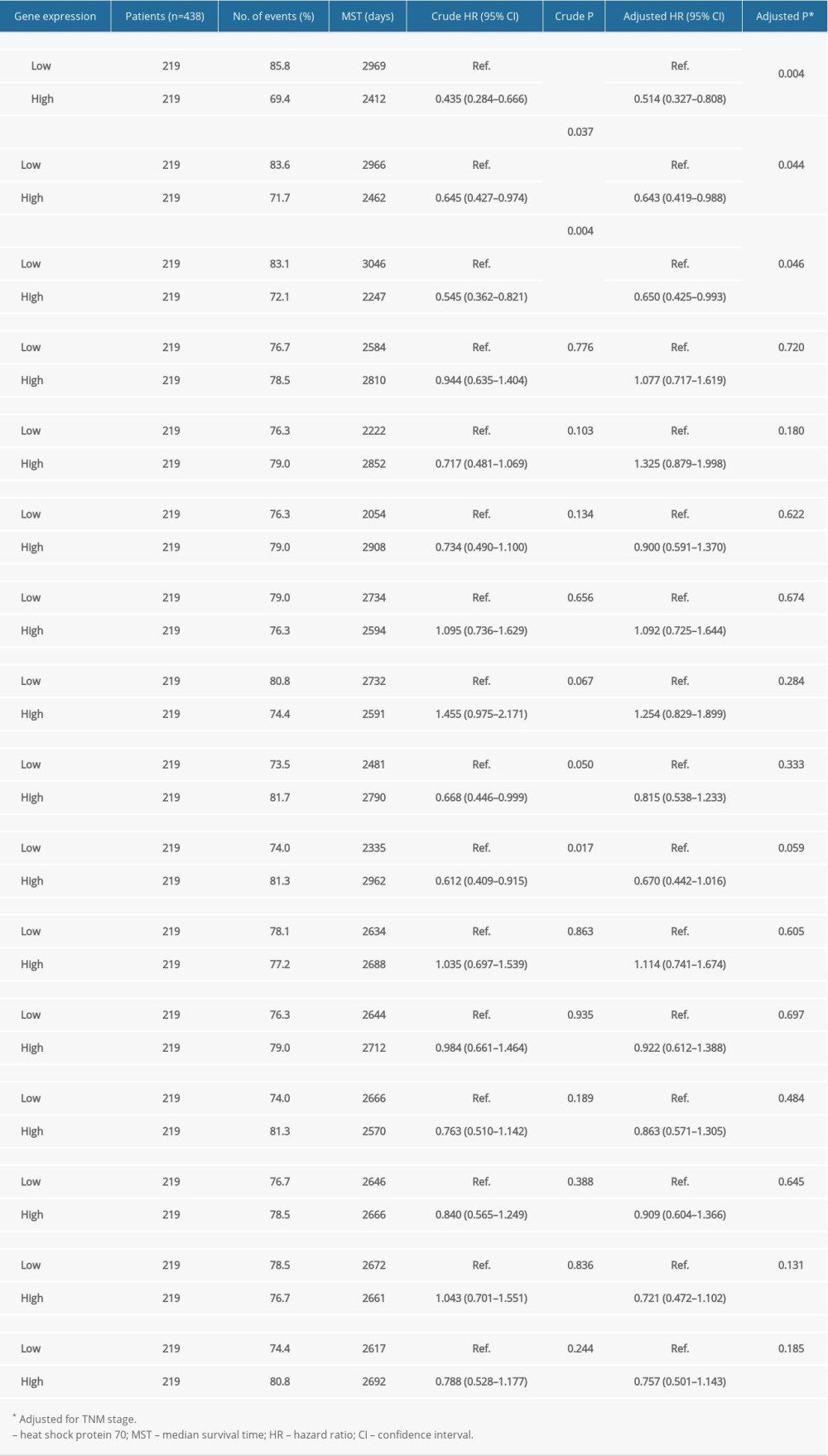 Table 2. Prognostic survival analysis of Hsp70 family genes.
Table 2. Prognostic survival analysis of Hsp70 family genes.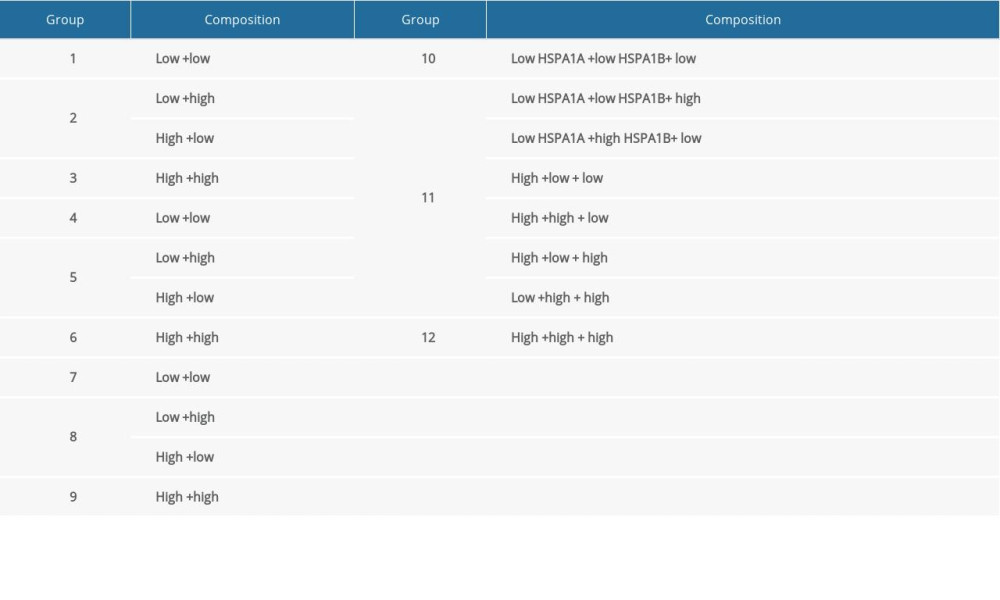 Table 3. Grouping according to HSPA1A, HSPA1B, and HSPA1L.
Table 3. Grouping according to HSPA1A, HSPA1B, and HSPA1L. Table 1. The clinical data for 438 patients with colorectal cancer.
Table 1. The clinical data for 438 patients with colorectal cancer. Table 2. Prognostic survival analysis of Hsp70 family genes.
Table 2. Prognostic survival analysis of Hsp70 family genes. Table 3. Grouping according to HSPA1A, HSPA1B, and HSPA1L.
Table 3. Grouping according to HSPA1A, HSPA1B, and HSPA1L. In Press
07 Mar 2024 : Clinical Research
Knowledge of and Attitudes Toward Clinical Trials: A Questionnaire-Based Study of 179 Male Third- and Fourt...Med Sci Monit In Press; DOI: 10.12659/MSM.943468
08 Mar 2024 : Animal Research
Modification of Experimental Model of Necrotizing Enterocolitis (NEC) in Rat Pups by Single Exposure to Hyp...Med Sci Monit In Press; DOI: 10.12659/MSM.943443
18 Apr 2024 : Clinical Research
Comparative Analysis of Open and Closed Sphincterotomy for the Treatment of Chronic Anal Fissure: Safety an...Med Sci Monit In Press; DOI: 10.12659/MSM.944127
08 Mar 2024 : Laboratory Research
Evaluation of Retentive Strength of 50 Endodontically-Treated Single-Rooted Mandibular Second Premolars Res...Med Sci Monit In Press; DOI: 10.12659/MSM.944110
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952








