19 August 2020: Database Analysis
Screening and Identification of Potential Peripheral Blood Biomarkers for Alzheimer’s Disease Based on Bioinformatics Analysis
Xin Wang1BCD, Lantao Wang1AEF*DOI: 10.12659/MSM.924263
Med Sci Monit 2020; 26:e924263
Abstract
BACKGROUND: Alzheimer’s disease (AD) is the leading cause of dementia worldwide; however, the molecular mechanisms underlying its pathogenesis remain unclear. The present study aimed to discover some potential peripheral blood biomarkers for early detection of patients with AD.
MATERIAL AND METHODS: Publicly available AD datasets – GSE18309 and GSE97760 – were obtained from the Gene Expression Omnibus database, and limma package from Bioconductor was employed to search for differently expressed genes (DEGs). Weighted correlation network analysis was performed to identify DEGs with highly synergistic changes, and functional annotation of DEGs was performed using gene set enrichment analysis and Metascape. STRING and Cytoscape were used to construct protein-protein interaction networks and analyze the most significant hub genes. Thereafter, the Comparative Toxicogenomics Database (CTD) was used to identify hub genes associated with AD pathology, and Connectivity Map was used to screen small molecule drugs for AD. Finally, hub genes coupled with corresponding predicted miRNAs involved in AD were assessed via TargetScan, and functional annotation of predicted miRNAs was performed using DIANA database.
RESULTS: Our analyses revealed 5042 DEGs; based on functional analyses, these DEGs were mainly associated with oligosaccharide lipid intermediate biosynthetic process, cyclin binding, signaling pathways regulating pluripotency of ubiquitin mediated proteolysis, and extracellular matrix-receptor interaction. UBB, UBA52, SRC, MMP9, VWF, GP6, and PF4 were identified as the hub genes. The CTD showed that these hub genes are closely related with AD or cognition impairment.
CONCLUSIONS: The identified hub genes and corresponding miRNAs might be useful as potential peripheral blood biomarkers of AD.
Keywords: Alzheimer Disease, Biological Markers, database, Computational Biology, Databases, Genetic, gene ontology, Gene Regulatory Networks, Protein Interaction Mapping
Background
Alzheimer’s disease (AD) is one of the most common neurodegenerative diseases and a major cause of dementia in elderly patients worldwide. Patients with AD often suffer from disorientation, language disorders, cognitive decline, and irreversible memory loss. The World Health Organization reported that the number of patients with AD is 50 million currently; it is projected to increase to 152 million by 2050 and, due to improved human longevity, eventually become the major cause of death in the world [1]. Significant global social and economic resources have been invested to clarify the molecular mechanisms underlying the pathogenesis of AD, but effective strategies for its prevention and treatment are still lacking [2,3]. Furthermore, at present, physicians diagnose AD based on their clinical experience and a series of complex examinations [4,5], and there is also a lack of simple and effective methods to diagnose early-stage AD. Thus, the increasing number of patients with AD and the consequent mounting social crisis has led physicians and biologists to invest tremendous amounts of time and effort in the discovery and development of potentially useful sensitive and precise biomarkers for AD in the cerebrospinal fluid (CSF) and blood [6]. Classical CSF biomarkers, such as amyloid-β and tau protein, were broadly studied as powerful and useful biomarkers for early diagnosis of AD. However, the failure of several clinical trials targeting amyloid-β and tau protein have proved that these CSF biomarkers are not reliable and suitable for monitoring dementia severity and predicting AD disease progression [7]. Moreover, CSF collection through lumbar puncture in the hospital setting does not seem to be a valid and minimally invasive population-wide screening tool [8]. Peripheral blood or plasma, on the other hand, can be collected relatively non-invasively and contain considerable disease-associated proteins. Therefore, peripheral blood or plasma can prove to be a source of potential genetic indicators for AD diagnosis and may pave the way for cost-effective AD risk screening in the middle-aged population prior to cognitive decline.
Extensive studies have been conducted to explore blood or plasma biomarkers for AD. In 2007, a genome-wide association study of 1086 controls and patients with sporadic AD revealed ApoE as a major risk gene for sporadic AD [9]. Ray et al. discovered 18 plasma signaling proteins which can be used to classify and predict the clinical Alzheimer’s diagnosis with approximate 90% accuracy [10]. Hadar et al. found 4 possible genes by comparing blood derived cell lines from AD patients and controls [11]. However, due to the variations and discrepancies in research methods, there are still no appropriate blood biomarkers for AD diagnosis.
Nowadays, microarray technology and bioinformatics analysis have been widely applied to identify differentially expressed genes (DEGs) and functional pathways in AD [12,13]. In this study, we identified 7 potential peripheral blood biomarkers for AD based on weighted correlation network analysis (WGCNA). The functional analysis suggests that the corresponding hub miRNAs may play a pivotal regulatory role in neuronal functions in AD; nevertheless, these results still need more experiments to investigate their validation and feasibility.
Material and Methods
MICROARRAY DATA:
The GSE18309 and GSE97760 gene expression profile datasets were obtained from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/), which is an open functional genomics database of high-throughput resources and includes microarray, gene expression and chips data [14]. Data for a total of 12 AD cases and 13 age-matched healthy controls were available (Table 1). The GSE18309 microarray dataset contains peripheral blood mononuclear cell transcriptomes from 3 patients with mild cognitive impairment, 3 patients with AD, as well as 3 normal controls. The GSE97760 microarray dataset contains blood RNA data from 9 women (age, 79.3±12.3 years) with advanced AD and 10 age–matched female healthy controls (age, 72.1±13.1 years) [15].
IDENTIFICATION OF DEGS:
For this study, the GSE18309 and GSE97760 datasets were merged, and data were normalized using R software. The DEGs were searched using the R package “limma” from (P-value, <0.05) [16].
WGCNA OF DEGS:
WGCNA can be used to explore and describe various combinations and associations of genes among different samples [17]. In this study, a total of 5042 DEGs were chosen for WGCNA, and gene sets with highly synergistic changes were searched. Thereafter, novel biomarkers were searched based on the correlations between gene sets and phenotypes.
FUNCTIONAL AND PATHWAY ENRICHMENT ANALYSIS:
Gene ontology (GO) is a knowledge base that is commonly used to search for functions of genes or individual genomic products; it considers 3 aspects: molecular functions (MF), biological processes (BP), and cellular components (CC) [18]. The Kyoto Encyclopedia of Genes and Genomes (KEGG) is a set of high-throughput gene and protein pathways [19]. Gene Set Enrichment Analysis (GSEA) is a computational method that assesses gene expression data and provides many biological pathways [20]. Metascape (http://metascape.org/gp/index.html) is a robust, publicly available database that facilitates comprehensive gene list annotation and analysis [21]. In this study, Metascape and GSEA analyses were employed to conduct GO and KEGG analyses for the DEGs (P<0.05).
PROTEIN–PROTEIN INTERACTION (PPI) NETWORKS:
STRING (
IDENTIFYING THE HUB GENES ASSOCIATED WITH AD:
The Comparative Toxicogenomics Database (CTD; http://ctdbase.org/) is a powerful open-access database for analyzing the associations between gene products and human diseases [22]. In our study, we made full use of this online database to unearth the connection between these identified hub genes and AD.
MIRNA PREDICTION OF HUB GENES AND FUNCTIONAL AND PATHWAY ENRICHMENT ANALYSIS:
TargetScan (www.targetscan.org) is an extensively approved and applied online database, which can effectively forecast the potential microRNA biological target sites [23]. In the present study, TargetScan was applied to find out some potential miRNAs that can regulate these identified hub genes. DIANA (DNA Intelligent Analysis) miRPath v3.0 is a popular online database used for functional and pathway enrichment analysis [24]. GO and KEGG pathway enrichment analysis for the predicted miRNAs were conducted using miRPath (P<0.05).
IDENTIFICATION OF SMALL MOLECULE DRUGS THAT REGULATED THE HUB GENES:
Connectivity Map (CMap) is a web-based database which connects human diseases with underlying genes and targeted drugs. In this study, we used the CMap database to identify small molecule drugs that regulated the genes associated with AD.
STATISTICAL ANALYSES:
In this study, all statistical analyses were conducted using SPSS 20.0 (Chicago, IL, USA), R software (version 3.5.0), and GraphPad Prism 7 (San Diego, CA, USA). The prediction efficiency was analyzed using the R package “survival ROC”. Univariate logistic proportional regression analysis was performed to analyze the effect of hub genes on AD.
Results
IDENTIFICATION OF DEGS:
In total, 5042 differentially expressed genes from the GSE18309 and GSE97760 datasets were distinguished in our study (Figure 1A). The corresponding DEG heatmaps are shown in Figure 1B.
WGCNA ANALYSIS OF DEGS:
The cluster of patients with clinical information is shown in Figure 1C. For this study, network topology analysis was used to identify the thresholding power as 12, and hierarchical clustering trees were produced based on this value. The ME-DissThres was set as 0.25 to merge similar modules and a total of 6 modules were generated (Figure 1D). The independence degrees among the 6 modules were explored using the DEG heatmap and dendrogram (Figure 1E, 1F, respectively), which demonstrate the high independence among modules. Yellow modules are most relevant to AD (Figure 1G).
FUNCTIONAL AND PATHWAY ENRICHMENT ANALYSIS:
Functional and pathway enrichment analysis of the DEGs in the yellow modules was performed using GSEA and Metascape. Based on GSEA, the results of GO analysis were found to be significantly enriched in oligosaccharide lipid intermediate biosynthetic process, nuclear localizing signal-bearing protein import into the nucleus, cyclin binding, neurotransmitter transporter activity, etc. (Figure 2A, Table 2). The results of KEGG analysis were significantly enriched in ubiquitin mediated proteolysis, cysteine and methionine metabolism, glycosphingolipid biosynthesis lacto and neolacto series, etc. (Figure 2B, Table 3). In Metascape, the enrichment results mainly involved mitophagy-animal, endosome membrane, platelet activation, etc. (Figure 2C–2E).
PPI NETWORK CONSTRUCTION AND IDENTIFICATION OF HUB GENES:
The PPI network of the DEGs was constructed using STRING and Cytoscape (Figure 3A). Four algorithms (Betweenness, Closeness, Radiality, and EcCentricity) were employed to search for hub genes (Figure 3B), which were the genes encoding ubiquitin B (UBB), ubiquitin A-52 residue ribosomal protein fusion product 1 (UBA52), SRC proto-oncogene, non-receptor tyrosine kinase (SRC), matrix metallopeptidase 9 (MMP9), von Willebrand factor (VWF), glycoprotein VI platelet (GP6), and platelet factor 4 (PF4) (Figure 3C, Table 4). The CTD database showed that the hub genes targeted AD related disease (Figure 3D).
HUB GENE MIRNA PREDICTION AND FUNCTIONAL AND PATHWAY ENRICHMENT ANALYSIS:
The miRNAs that regulate the hub genes are shown in Table 5. Functional and pathway enrichment analysis of the miRNAs is shown in Figure 4. The results of GO analysis using the DIANA-miRPath software showed that the variations in the “BP” of miRNAs which regulate hub genes were mainly enriched in organelle, ion binding, cellular protein modification process, etc. (Figure 4A). The “CC” items of the miRNAs were significantly enriched in cellular component assembly, cell junction organization, small molecule metabolic process, etc. (Figure 4B). As for the “MF”, miRNAs which regulate hub genes were observably enriched in molecular function, Fc-epsilon receptor, cytosol, etc. (Figure 4C). According to the results of KEGG pathway analysis, the top meaningful pathways associated with miRNAs which regulate hub genes were TGF-β signaling pathway, MAPK signaling pathway, neurotrophin signaling pathway, etc. (Figure 4D).
IDENTIFICATION OF CANDIDATE SMALL MOLECULE DRUGS FOR TREATING AD:
In this study, CMap database analysis was employed to explore potential small molecule drugs for AD. The cutoff criteria were number of instances >2 and P-value <0.05. The identified molecules are listed in Table 6.
PREDICTION EFFICIENCY AND EFFECT OF HUB GENES:
Receiver operating characteristic (ROC) curve analysis was performed to assess the predictive value of the hub genes (Figure 5). UBA52 and UBB had the highest areas under the curves. The odd ratios and 95% confidence intervals in univariate analysis for AD are shown in Table 7.
Discussion
It is known that AD is the main cause of dementia among elderly populations and that it is characterized by the excessive deposition of extracellular plaque formed by amyloid protein, intracellular neurofibrillary tangles formed by phosphorylated tau protein, and selective loss of susceptible neurons. However, the exact mechanisms underlying the pathogenesis of AD have not been clarified, and treatment options are limited. The increasing prevalence of AD, along with improved human longevity, imposes immense economic and social burdens on the world. The only sensible solution to this crisis would be the development of effective and affordable diagnostics for AD detection so that the progression of AD can be addressed or halted prior to dementia onset. Hence, it is of vital importance to explore and develop more affordable, convenient, and efficient techniques and peripheral blood biomarkers for the detection and diagnosis of AD [8]. In this study, we searched for potential miRNA biomarkers in the peripheral blood of patients with AD based on microarray technology and bioinformatic analyses, which may advance our understanding of AD and then contribute to bring out more comprehensive, effective and feasible prevention strategies.
The recent great advance and widely application of bioinformatics technologies have promoted to explore more feasible biomarkers and more novel therapeutic candidates in various diseases. More and more sequences data are being submitted to public databases such as The Cancer Genome Atlas and GEO databases. In this study, transcriptome expression profiles of patients with AD were downloaded from the GEO database and re-analyzed to search for potential peripheral blood biomarkers of AD; using bioinformatics analyses, we found 5042 DEGs between AD and age-matched normal samples. GO and KEGG analyses of these DEGs were carried out to investigate the potential underlying molecular mechanisms and connections between them. It is worth noting that “mitophagy” was enriched; mitochondrial dysfunction contributes to a wide spectrum of age-related diseases such as AD, Parkinson’s disease, and other neurodegenerative diseases. Emerging findings demonstrate that mitochondrial dysfunction, accumulation of damaged mitochondria, and impaired mitophagy occurs early in neurons affected by AD, which could promote the disease-defining amyloid-β peptide and tau pathologies [25–27]. Thus, bolstering mitochondrial health and/or stimulating mitophagy may forestall the neurodegenerative process in AD. Moreover, GO analysis based on GSEA indicated enrichment in “neurotransmitter transporter activity”. Acetylcholine is one of the most important neurotransmitters that have an important in the central nervous systems. It has been reported that loss of cholinergic neurons contributes to memory and attention deficits, which could lead to disease progression and clinical condition aggravation [28]. Cholinesterase inhibitor therapy has been developed as the standard and predominant treatment for relieving symptoms and possibly slowing down disease progression of AD [29,30]. Thus, drugs that act on the neurotransmitter transporter may represent a promising option to relieve and cure patients with AD.
In this study, 7 hub genes (UBB, UBA52, SRC, MMP9, VWF, GP6, and PF4) were determined based on PPI network analysis. UBB and UBA52 encode ubiquitin, which plays a key role in 26S proteasomal degradation of target cell proteins. An abnormal form of ubiquitin was found to accumulate in the brain tissues of patients with AD; meanwhile APP+1 and UBB+1 frameshift proteins were demonstrated to elicit the production of AD-specific autoantibodies [31–33]. SRC encodes a tyrosine-protein kinase that is associated with cell growth. Scales et al. have demonstrated that the SRC family plays a key role in hyper-phosphorylation of tau protein, which is the main component of neurofibrillary tangles in AD [34]. Moreover, inhibition of SRC family kinases can improve spatial cognitive function in a non-traumatic intraventricular hemorrhage rat model [35]. Therefore, inhibiting SRC expression may also affect the pathology and severity of AD. MMP9 is involved in the breakdown of the extracellular matrix, tissue remodeling, cellular receptor stripping, and processing of various signaling molecules. It has been shown that the potential of MMP family proteins to degrade amyloid-β is compromised in the brains of patients with AD [36]. Thus, MMP9 hypofunction in the brain may contribute to the anomalous deposits associated with AD. VWF encodes a glycoprotein involved in hemostasis. Vascular damage was thought to play a key role in the pathogenesis of AD, and VWF has also been considered as a biomarker for vascular damage [37,38]. Moreover, patients with AD have an increased plasma level of VWF [39], indicating that it could be used as an early biomarker of AD. GP6 (glycoprotein VI platelet), a platelet membrane glycoprotein of the immunoglobulin superfamily, plays a critical role in collagen-induced platelet aggregation and thrombus formation. A growing number of evidence from clinical and basic studies indicates that vascular risk factors and angiopathic mechanisms are extensively involved in the pathogenesis of AD. GP6 plasma levels are significantly decreased in patients with AD compared with those in healthy controls, and GP6 levels are also positively correlated with Mini-Mental State Examination score [40]. Thus, we hypothesize that GP6 plays a neuroprotective role in the pathogenesis of AD. PF4 encodes a member of the CXC chemokine family. It has been reported that, in the presence of Aβ42, uncontrolled activation of platelets could lead to a chronic inflammatory reaction due to the over-secretion of PF4 [41]. Thus, targeting PF4 may provide a new avenue for anti-AD therapy. Our results from the CTD demonstrate that all hub genes are involved in neurodegenerative disease, cognitive disorder, memory disorder, or learning disorder. Moreover, our ROC analysis results showed that the predictive value of the hub genes were relatively high. All these evidences support the possibility that these hub genes may be potential peripheral blood biomarkers for AD diagnosis; however, more
Recent accumulating evidences show that miRNAs may be an attractive type of blood-based biomarkers for patients with AD because miRNAs are relatively more stable in serum and plasma than mRNAs. In our study, the comprehensive functional and pathway enrichment analysis results of miRNAs made it clear that they may play an essential role in the occurrence and development of AD. Interestingly, alteration of hsa-miR-9-5p has been demonstrated to be closely related with AD pathology [42,43]. All these data acquired through bioinformatics analyses supply valuable information about how these predicted miRNAs are related to the AD progression.
Conclusions
In summary, our work to identify peripheral blood biomarkers for patients with AD provides some valuable potential biological targets for further and deeper mechanistic investigation, which could vigorously accelerate the pace to develop more simple, effective and feasible screening methods for populations at high risk of AD.
Figures
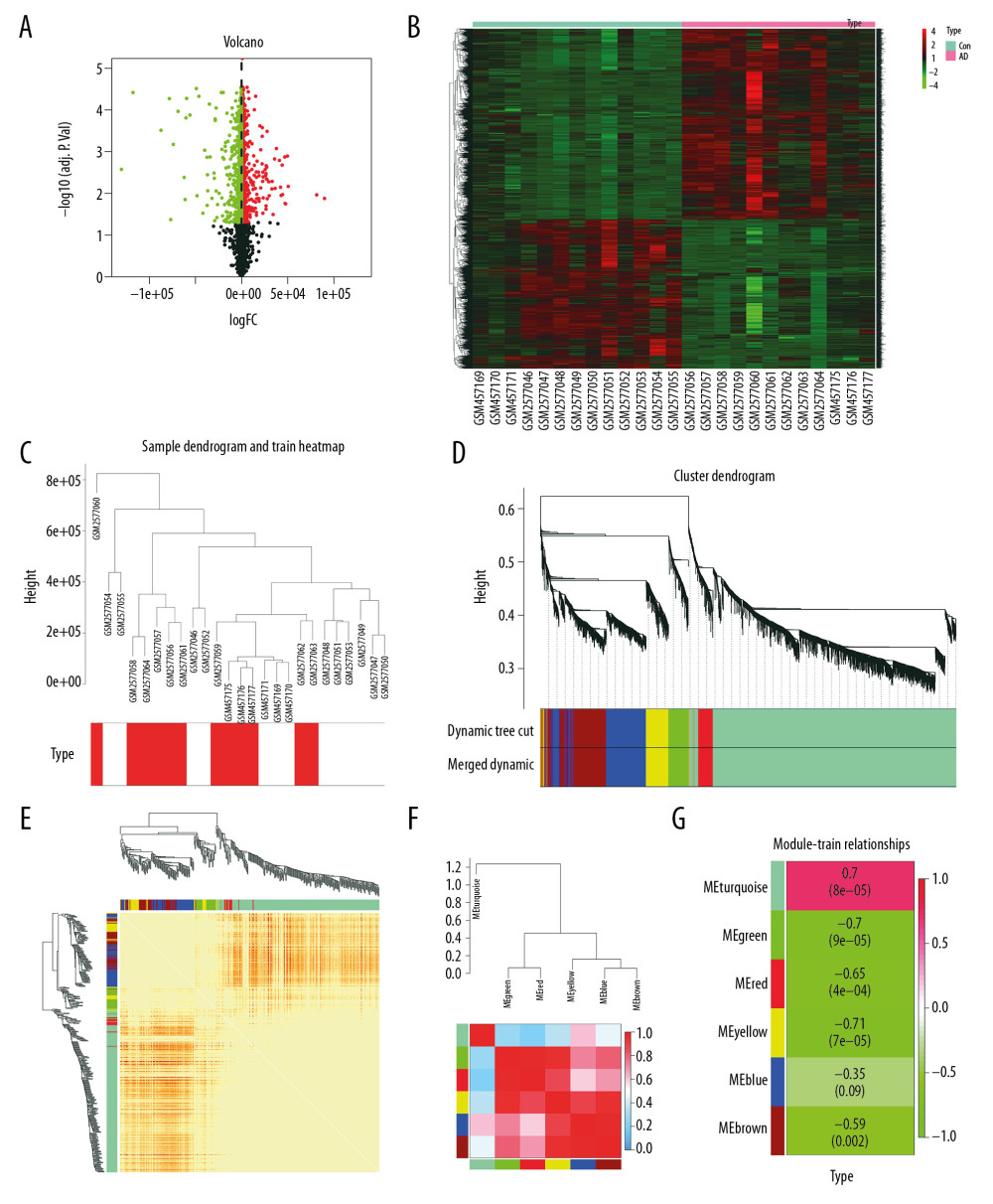 Figure 1. Differential expression analysis and WGCNA analysis of the genes in the merged series. (A) Volcano plots of the DEGs between AD group and control group. (B) Heatmaps of the DEGs in the merged series. (C) Cluster of patients with clinical information; the red line represents patients with AD. (D) Repeated hierarchical clustering tree of the 5042 genes. (E) Dendrogram and heatmap of the DEGs. (F) Interactions between these modules. (G) Associations between clinical traits and the modules; the MEyellow module has the highest correlation with clinical traits. WGCNA – weighted correlation network analysis; DEGs – differently expressed genes; AD – Alzheimer’s disease.
Figure 1. Differential expression analysis and WGCNA analysis of the genes in the merged series. (A) Volcano plots of the DEGs between AD group and control group. (B) Heatmaps of the DEGs in the merged series. (C) Cluster of patients with clinical information; the red line represents patients with AD. (D) Repeated hierarchical clustering tree of the 5042 genes. (E) Dendrogram and heatmap of the DEGs. (F) Interactions between these modules. (G) Associations between clinical traits and the modules; the MEyellow module has the highest correlation with clinical traits. WGCNA – weighted correlation network analysis; DEGs – differently expressed genes; AD – Alzheimer’s disease. 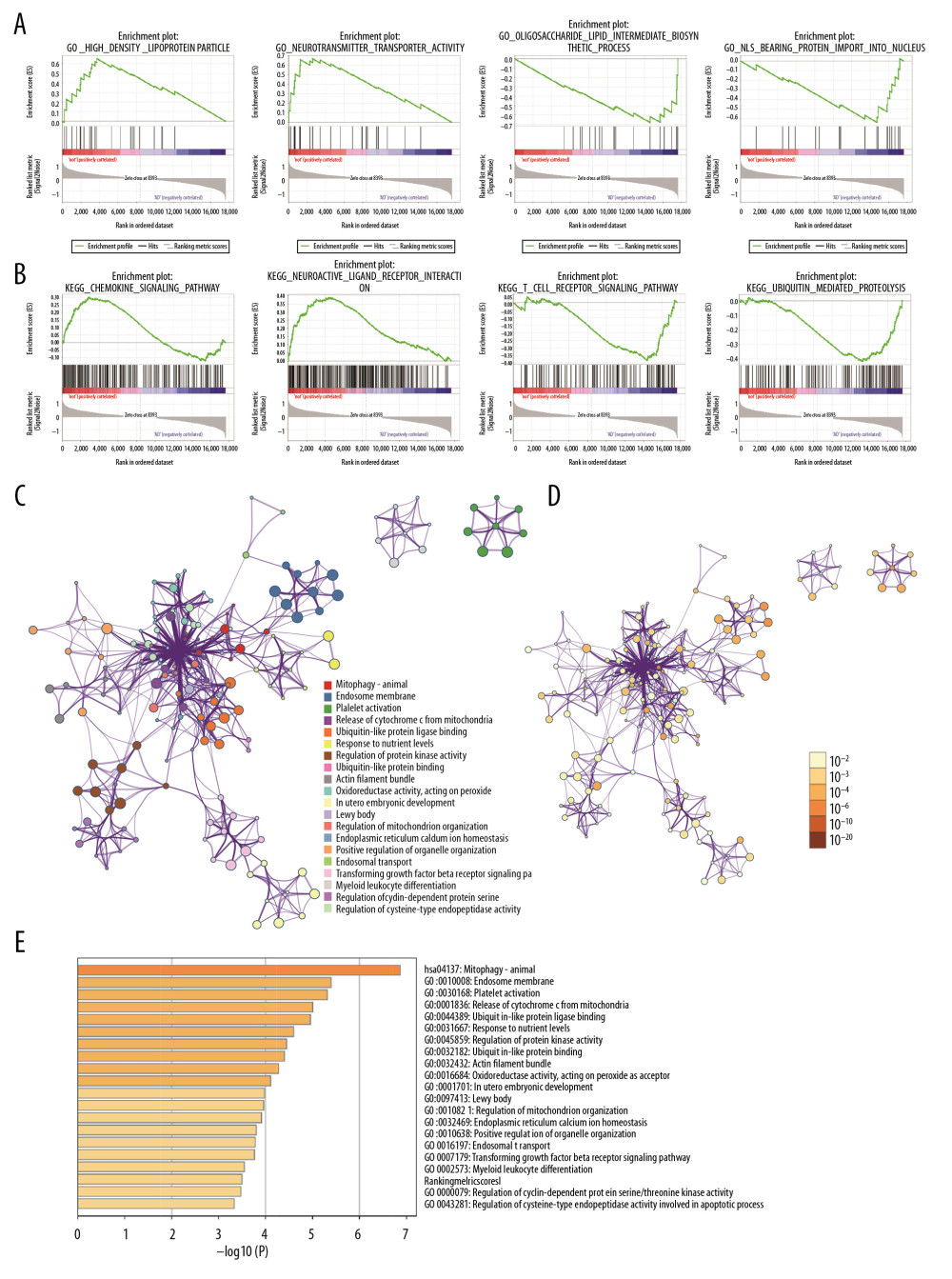 Figure 2. Gene functional enrichment analysis of the MEyellow model DEGs based on GSEA and Metascape. (A) GSEA-based GO analyses. (B) GSEA-based KEGG analyses. (C) Enrichment GO-KEGG color by cluster analyses using Metascape. (D) Enrichment GO-KEGG Color by P-value analyses using Metascape. (E) Enrichment heatmap selected GO-KEGG analyses using Metascape. DEGs – differently expressed genes; GSEA – Gene Set Enrichment Analysis; GO – Gene Ontology; KEGG – Kyoto Encyclopedia of Genes and Genomes.
Figure 2. Gene functional enrichment analysis of the MEyellow model DEGs based on GSEA and Metascape. (A) GSEA-based GO analyses. (B) GSEA-based KEGG analyses. (C) Enrichment GO-KEGG color by cluster analyses using Metascape. (D) Enrichment GO-KEGG Color by P-value analyses using Metascape. (E) Enrichment heatmap selected GO-KEGG analyses using Metascape. DEGs – differently expressed genes; GSEA – Gene Set Enrichment Analysis; GO – Gene Ontology; KEGG – Kyoto Encyclopedia of Genes and Genomes. 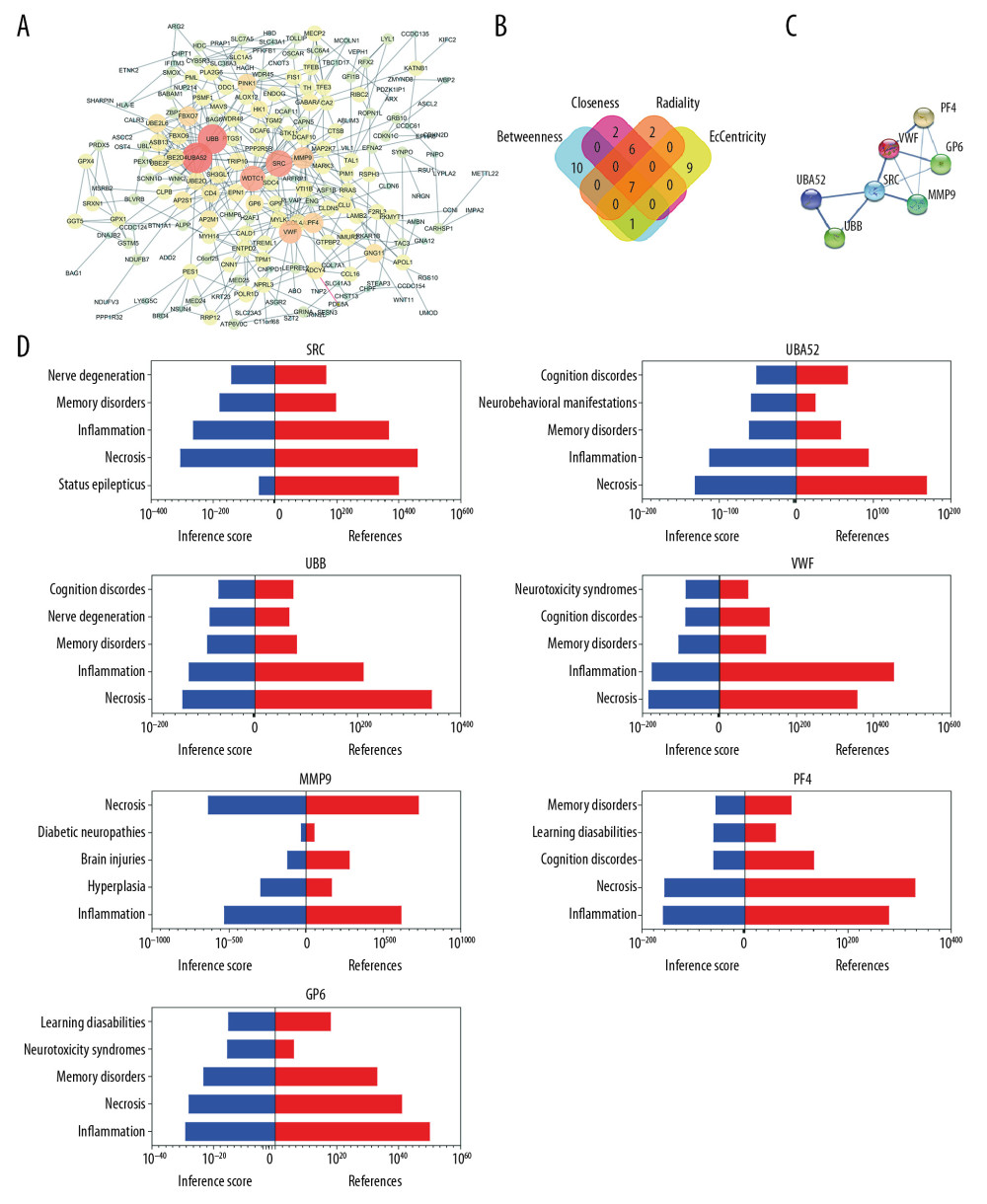 Figure 3. Relationship between DEGs. (A) The PPI network; the more the number of connections, the more likely the interaction. (B) Common hub genes identified using different algorithms. (C) Common hub genes of the PPI network. (D) Relationship to AD group and control group related to hub genes based on the CTD. DEGs – differently expressed genes; PPI – protein–protein interactions; AD – Alzheimer’s disease; CTD – Comparative Toxicogenomics Database.
Figure 3. Relationship between DEGs. (A) The PPI network; the more the number of connections, the more likely the interaction. (B) Common hub genes identified using different algorithms. (C) Common hub genes of the PPI network. (D) Relationship to AD group and control group related to hub genes based on the CTD. DEGs – differently expressed genes; PPI – protein–protein interactions; AD – Alzheimer’s disease; CTD – Comparative Toxicogenomics Database. 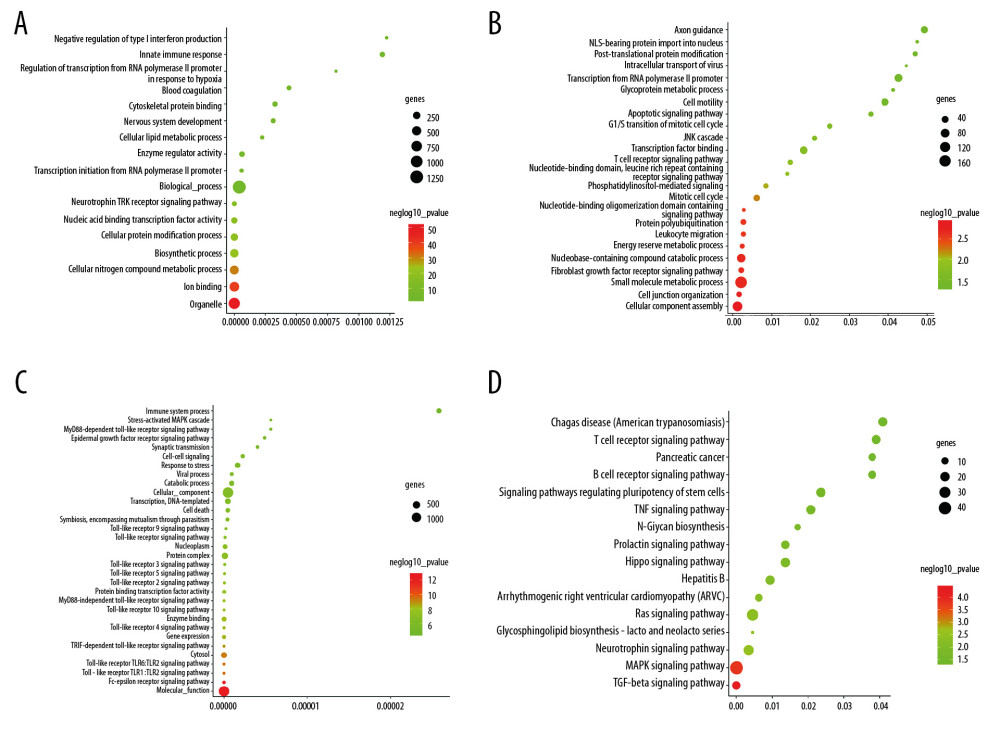 Figure 4. Functional and pathway enrichment analyses of miRNAs which could regulate hub genes. (A) BP analyses. (B) CC analyses. (C) MF analyses. (D) KEGG analyses. miRNAs – microRNAS; BP – biological processes; CC – cellular components; MF – molecular functions; KEGG – Kyoto Encyclopedia of Genes and Genomes.
Figure 4. Functional and pathway enrichment analyses of miRNAs which could regulate hub genes. (A) BP analyses. (B) CC analyses. (C) MF analyses. (D) KEGG analyses. miRNAs – microRNAS; BP – biological processes; CC – cellular components; MF – molecular functions; KEGG – Kyoto Encyclopedia of Genes and Genomes. 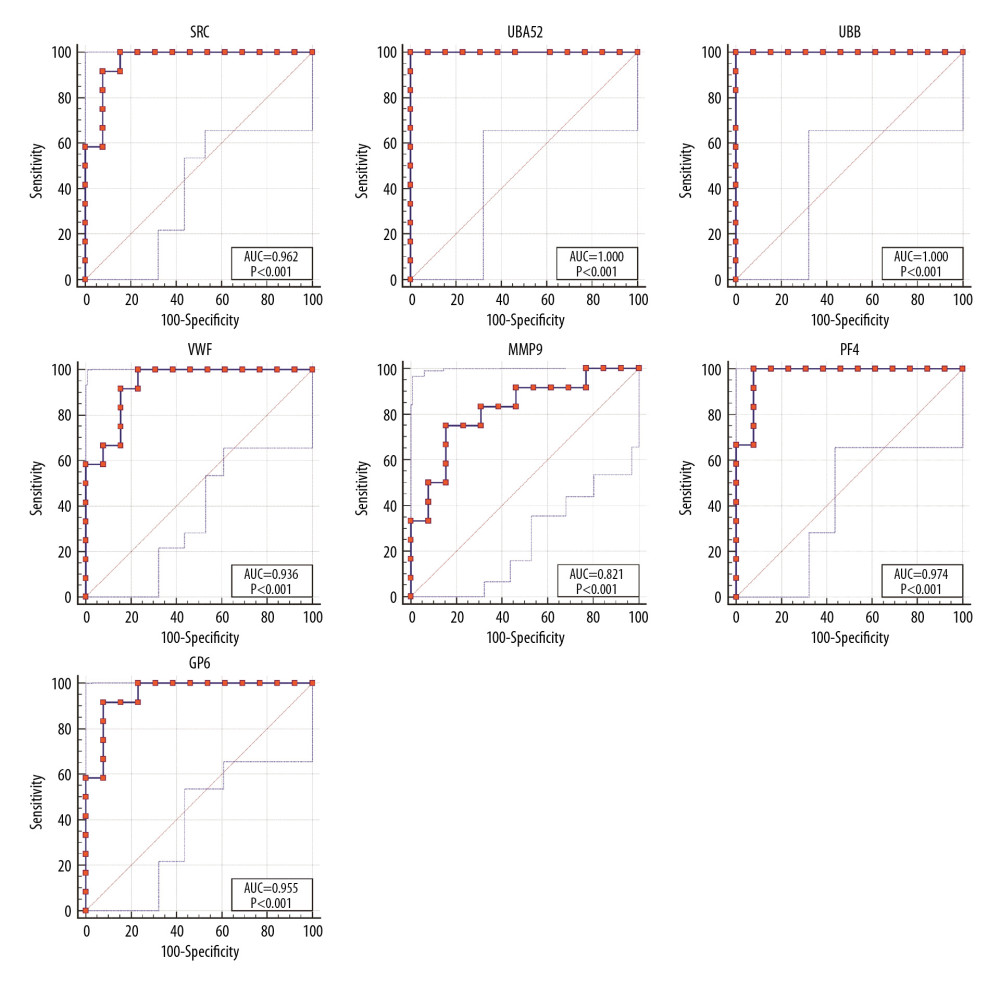 Figure 5. ROC curves of the hub genes. ROC – receiver operating characteristic.
Figure 5. ROC curves of the hub genes. ROC – receiver operating characteristic. Tables
Table 1. Summary of Alzheimer’s disease (AD) microarray datasets from different Gene Expression Omnibus (GEO) datasets.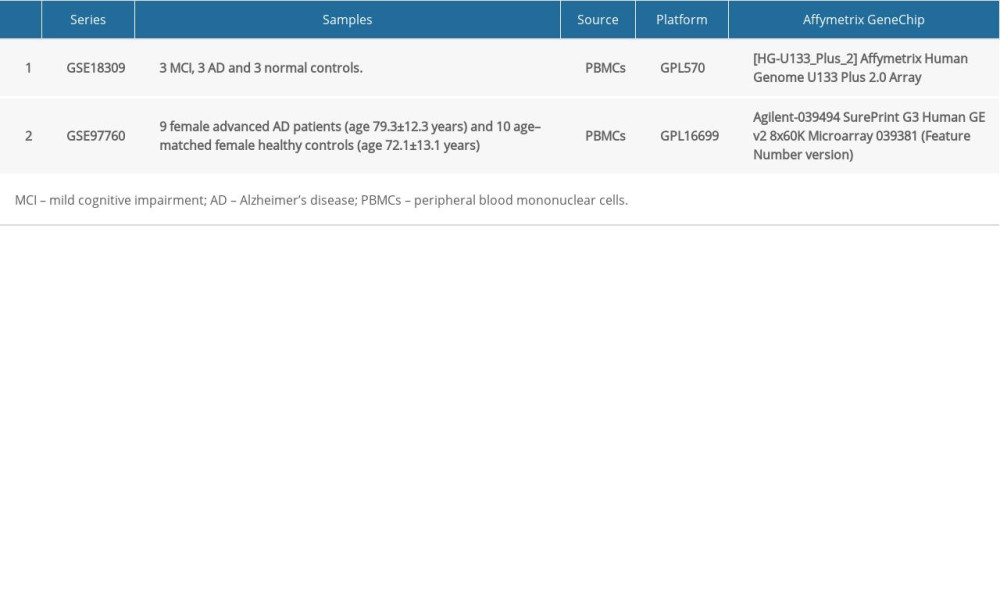 Table 2. Gene Set Enrichment Analysis (GSEA)-based Gene Ontology (GO) snalysis.
Table 2. Gene Set Enrichment Analysis (GSEA)-based Gene Ontology (GO) snalysis.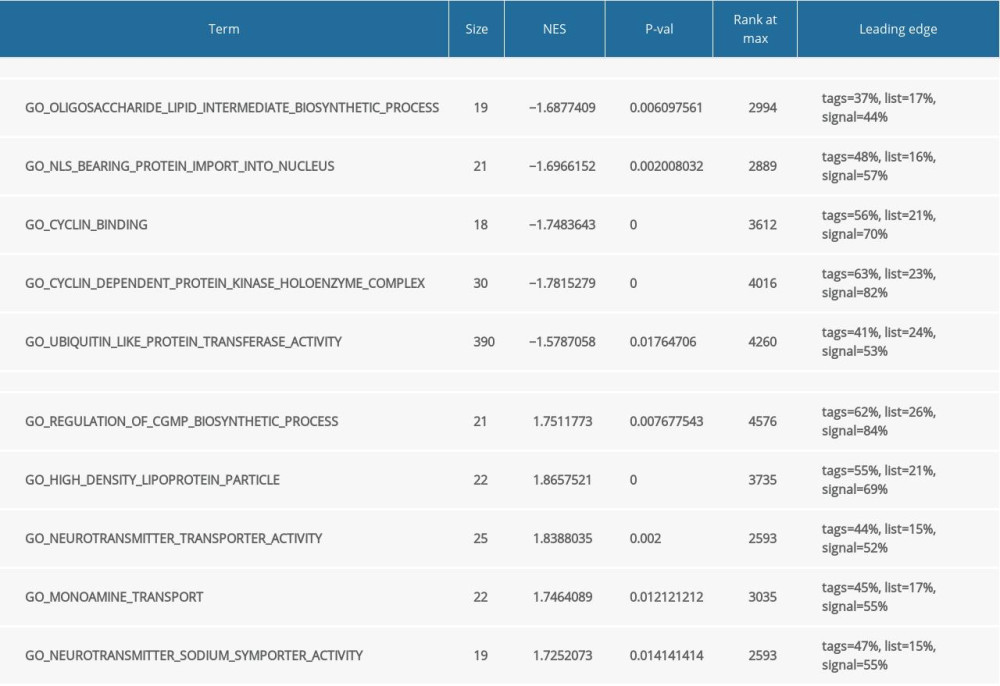 Table 3. Gene Set Enrichment Analysis (GSEA)-based and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis.
Table 3. Gene Set Enrichment Analysis (GSEA)-based and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis.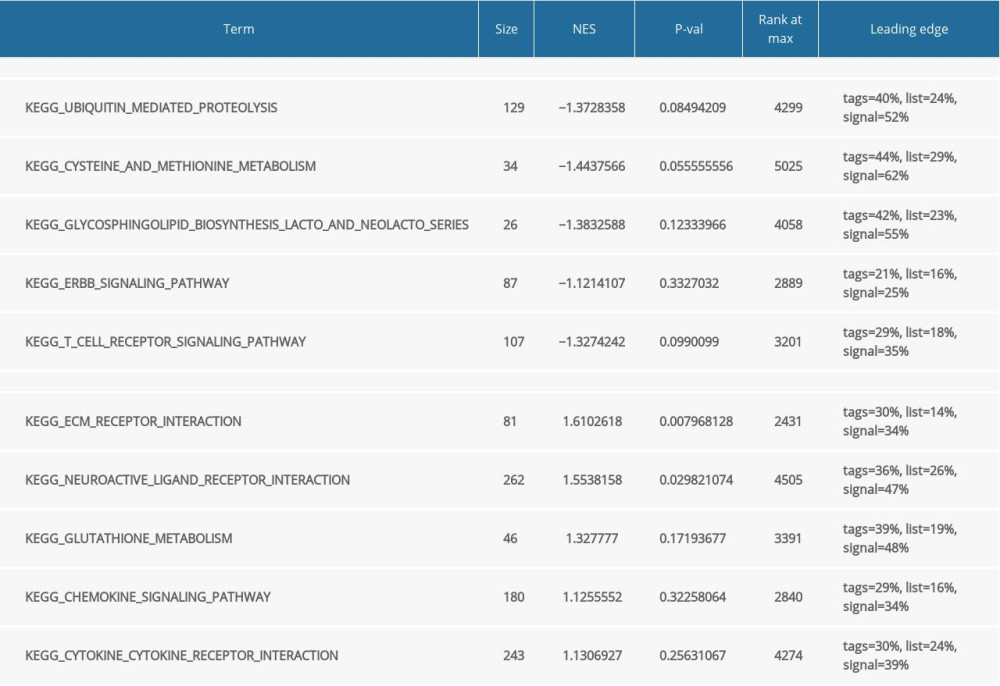 Table 4. Summary of hub genes.
Table 4. Summary of hub genes.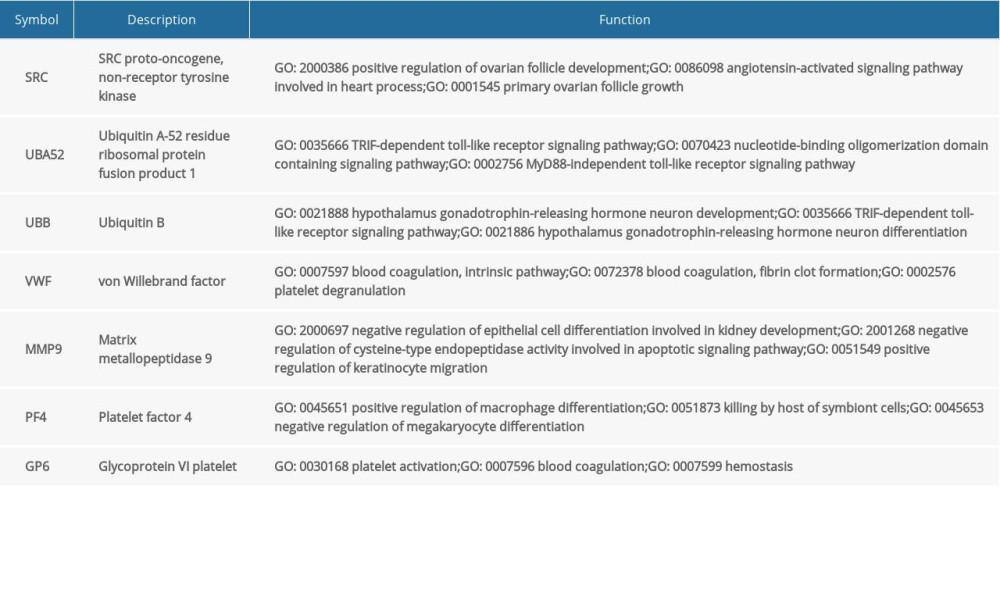 Table 5. Summary of MiRNAs that regulate hub genes.
Table 5. Summary of MiRNAs that regulate hub genes.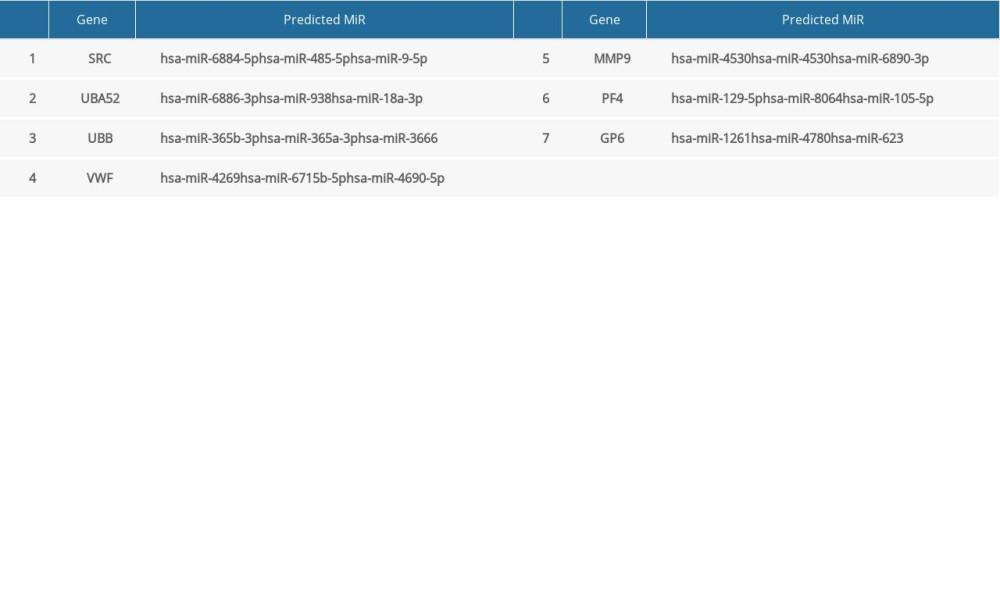 Table 6. Summary of hub gene regulation connectivity map.
Table 6. Summary of hub gene regulation connectivity map.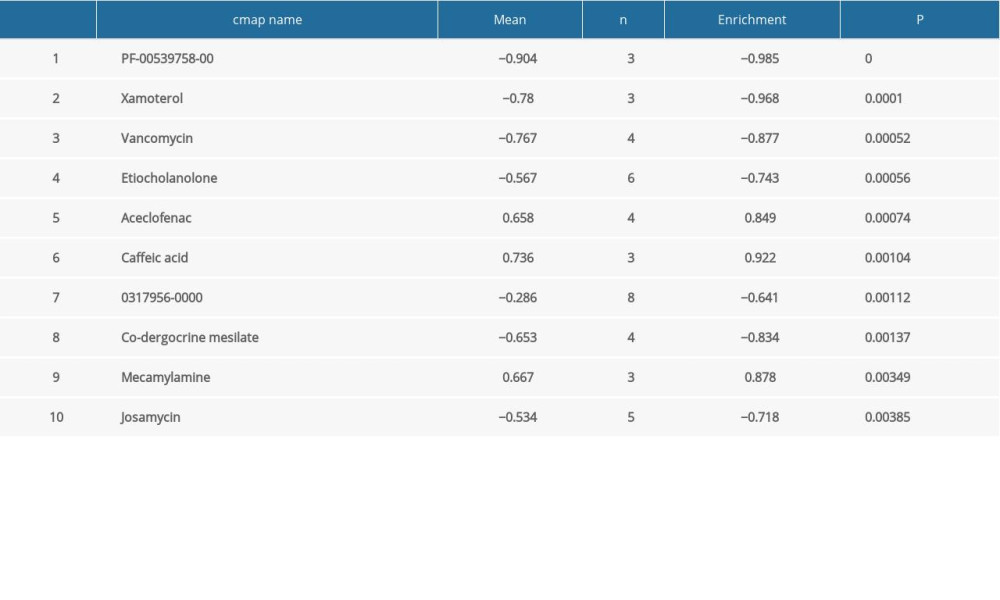 Table 7. Hub genes and their predicted effects on Alzheimer’s disease based on univariate logistic proportional regression analysis.
Table 7. Hub genes and their predicted effects on Alzheimer’s disease based on univariate logistic proportional regression analysis.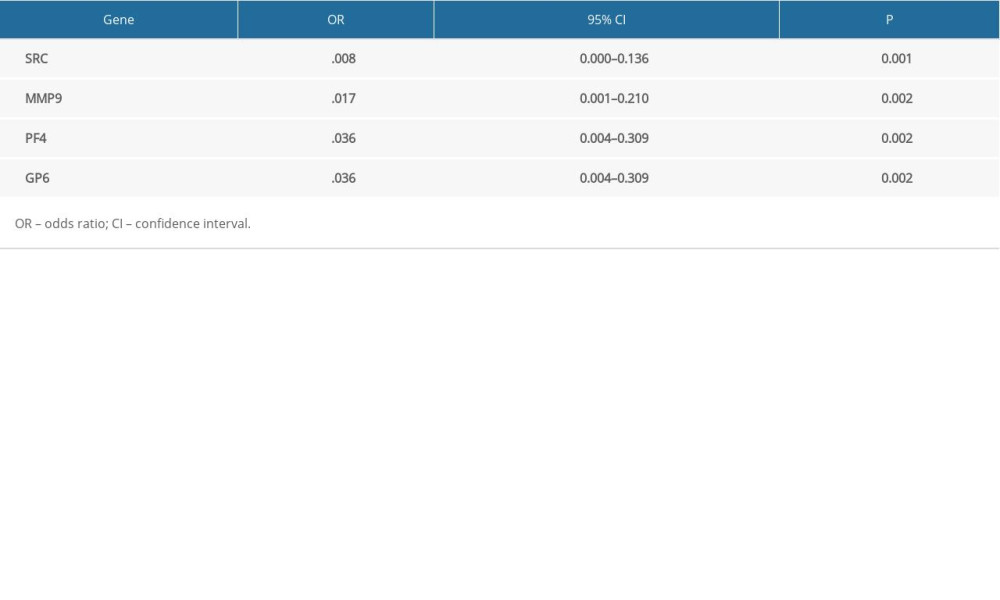
References
1. Hodson R, Alzheimer’s disease: Nature, 2018; 559(7715); S1
2. Fessel J, Alzheimer’s disease combination treatment: Neurobiol Aging, 2018; 63; 165
3. Atkinson SP, Alzheimer’s disease: A special collection: Stem Cells Transl Med, 2017; 6(11); 1951-55
4. Karlawish J, Jack CR, Rocca WA, Alzheimer’s disease: The next frontier – Special Report 2017: Alzheimers Dement, 2017; 13(4); 374-80
5. Sochocka M, Zwolinska K, Leszek J, The infectious etiology of Alzheimer’s disease: Curr Neuropharmacol, 2017; 15(7); 996-1009
6. Mantzavinos V, Alexiou A, Biomarkers for Alzheimer’s disease diagnosis: Curr Alzheimer Res, 2017; 14(11); 1149-54
7. Park SA, Han SM, Kim CE, New fluid biomarkers tracking non-amyloid-beta and non-tau pathology in Alzheimer’s disease: Exp Mol Med, 2020; 52(4); 556-68
8. Hadar A, Gurwitz D, Peripheral transcriptomic biomarkers for early detection of sporadic Alzheimer disease?: Dialogues Clin Neurosci, 2018; 20(4); 293-300
9. Villain N, Dubois B, Alzheimer’s disease including focal presentations: Semin Neurol, 2019; 39(2); 213-26
10. Ray S, Britschgi M, Herbert C, Classification and prediction of clinical Alzheimer’s diagnosis based on plasma signaling proteins: Nat Med, 2007; 13(11); 1359-62
11. Hadar A, Milanesi E, Squassina A, RGS2 expression predicts amyloid-β sensitivity, MCI and Alzheimer’s disease: Genome-wide transcriptomic profiling and bioinformatics data mining: Transl Psychiatry, 2016; 6(10); e909
12. Agyei D, Tsopmo A, Udenigwe CC, Bioinformatics and peptidomics approaches to the discovery and analysis of food-derived bioactive peptides: Anal Bioanal Chem, 2018; 410(15); 3463-72
13. Huang X, Liu S, Wu L, High throughput single cell RNA sequencing, bioinformatics analysis and applications: Adv Exp Med Biol, 2018; 1068; 33-43
14. Clough E, Barrett T, The Gene Expression Omnibus database: Methods Mol Biol, 2016; 1418; 93-110
15. Naughton BJ, Duncan FJ, Murrey DA, Blood genome-wide transcriptional profiles reflect broad molecular impairments and strong blood-brain links in Alzheimer’s disease: J Alzheimers Dis, 2015; 43(1); 93-108
16. Smyth GK, limma: Linear models for microarray data: Bioinformatics and computational biology solutions using R and bioconductor, 2005; 397-420, Springer
17. Langfelder P, Horvath S, WGCNA: An R package for weighted correlation network analysis: BMC Bioinformatics, 2008; 9; 559
18. Ashburner M, Ball CA, Blake JA, Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium: Nat Genet, 2000; 25(1); 25-29
19. Kanehisa M, The KEGG database: Novartis Foundation Symposium, 2002; 247; 91-101 discussion 101–103, 119–28, 244–52
20. Subramanian A, Tamayo P, Mootha VK, Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles: Proc Natl Acad Sci USA, 2005; 102(43); 15545-50
21. Zhou Y, Zhou B, Pache L, Metascape provides a biologist-oriented resource for the analysis of systems-level datasets: Nat Commun, 2019; 10(1); 1523
22. Davis AP, Grondin CJ, Johnson RJ, The Comparative Toxicogenomics Database: Update 2019: Nucleic Acids Res, 2019; 47(D1); D948-54
23. Agarwal V, Bell GW, Nam JW, Bartel DP, Predicting effective microRNA target sites in mammalian mRNAs: eLife, 2015; 4; e05005
24. Vlachos IS, Zagganas K, Paraskevopoulou MD, DIANA-miRPath v3.0: deciphering microRNA function with experimental support: Nucleic Acids Res, 2015; 43(W1); W460-66
25. Kerr JS, Adriaanse BA, Greig NH, Mitophagy and Alzheimer’s disease: Cellular and molecular mechanisms: Trends Neurosci, 2017; 40(3); 151-66
26. Fivenson EM, Lautrup S, Sun N, Mitophagy in neurodegeneration and aging: Neurochem Int, 2017; 109; 202-9
27. Fang EF, Hou Y, Palikaras K, Mitophagy inhibits amyloid-beta and tau pathology and reverses cognitive deficits in models of Alzheimer’s disease: Nat Neurosci, 2019; 22(3); 401-12
28. Ferreira-Vieira TH, Guimaraes IM, Silva FR, Ribeiro FM, Alzheimer’s disease: Targeting the cholinergic system: Curr Neuropharmacol, 2016; 14(1); 101-15
29. Sharma K, Cholinesterase inhibitors as Alzheimer’s therapeutics (eeview): Mol Med Rep, 2019; 20(2); 1479-87
30. Haake A, Nguyen K, Friedman L, An update on the utility and safety of cholinesterase inhibitors for the treatment of Alzheimer’s disease: Expert Opin Drug Saf, 2020; 19(2); 147-57
31. Munari F, Bortot A, Assfalg M, D’Onofrio M, Alzheimer’s disease-associated ubiquitin mutant Ubb(+1): Properties of the carboxy-terminal domain and its influence on biomolecular interactions: Int J Biol Macromol, 2018; 108; 24-31
32. Montero-Calle A, San Segundo-Acosta P, Garranzo-Asensio M, The molecular misreading of APP and UBB induces a humoral immune response in Alzheimer’s disease patients with diagnostic ability: Mol Neurobiol, 2020; 57(2); 1009-20
33. Seynnaeve D, Vecchio MD, Fruhmann G, Recent insights on Alzheimer’s disease originating from yeast models: Int J Mol Sci, 2018; 19(7); 1947
34. Scales TM, Derkinderen P, Leung KY, Tyrosine phosphorylation of tau by the SRC family kinases lck and fyn: Mol Neurodegener, 2011; 6; 12
35. Liu DZ, Waldau B, Ander BP, Inhibition of SRC family kinases improves cognitive function after intraventricular hemorrhage or intraventricular thrombin: J Cereb Blood Flow Metab, 2017; 37(7); 2359-67
36. Hussain AA, Lee Y, Zhang JJ, Disturbed matrix metalloproteinase pathway in both age-related macular degeneration and Alzheimer’s disease: J Neurodegener Dis, 2017; 2017 4810232
37. Thomas T, Miners S, Love S, Post-mortem assessment of hypoperfusion of cerebral cortex in Alzheimer’s disease and vascular dementia: Brain, 2015; 138(Pt 4); 1059-69
38. Yavuz BB, Dede DS, Yavuz B, Potential biomarkers for vascular damage in Alzheimer’s disease: Thrombomodulin and von Willebrand factor: J Nutr Health Aging, 2010; 14(6); 439-41
39. de Mello Gomide Loures C, Figueiredo Duarte RC, Ferreira Silva MV, Hemostatic abnormalities in dementia: A systematic review and meta-analysis: Semin Thromb Hemost, 2019; 45(5); 514-22
40. Laske C, Leyhe T, Stransky E, Association of platelet-derived soluble glycoprotein VI in plasma with Alzheimer’s disease: J Psychiatr Res, 2008; 42(9); 746-51
41. Zhang W, Huang W, Jing F, Contribution of blood platelets to vascular pathology in Alzheimer’s disease: J Blood Med, 2013; 4; 141-47
42. Riancho J, Vazquez-Higuera JL, Pozueta A, MicroRNA Profile in patients with Alzheimer’s disease: analysis of miR-9-5p and miR-598 in raw and exosome enriched cerebrospinal fluid samples: J Alzheimers Dis, 2017; 57(2); 483-91
43. Kiko T, Nakagawa K, Tsuduki T, MicroRNAs in plasma and cerebrospinal fluid as potential markers for Alzheimer’s disease: J Alzheimers Dis, 2014; 39(2); 253-59
Figures
 Figure 1. Differential expression analysis and WGCNA analysis of the genes in the merged series. (A) Volcano plots of the DEGs between AD group and control group. (B) Heatmaps of the DEGs in the merged series. (C) Cluster of patients with clinical information; the red line represents patients with AD. (D) Repeated hierarchical clustering tree of the 5042 genes. (E) Dendrogram and heatmap of the DEGs. (F) Interactions between these modules. (G) Associations between clinical traits and the modules; the MEyellow module has the highest correlation with clinical traits. WGCNA – weighted correlation network analysis; DEGs – differently expressed genes; AD – Alzheimer’s disease.
Figure 1. Differential expression analysis and WGCNA analysis of the genes in the merged series. (A) Volcano plots of the DEGs between AD group and control group. (B) Heatmaps of the DEGs in the merged series. (C) Cluster of patients with clinical information; the red line represents patients with AD. (D) Repeated hierarchical clustering tree of the 5042 genes. (E) Dendrogram and heatmap of the DEGs. (F) Interactions between these modules. (G) Associations between clinical traits and the modules; the MEyellow module has the highest correlation with clinical traits. WGCNA – weighted correlation network analysis; DEGs – differently expressed genes; AD – Alzheimer’s disease. Figure 2. Gene functional enrichment analysis of the MEyellow model DEGs based on GSEA and Metascape. (A) GSEA-based GO analyses. (B) GSEA-based KEGG analyses. (C) Enrichment GO-KEGG color by cluster analyses using Metascape. (D) Enrichment GO-KEGG Color by P-value analyses using Metascape. (E) Enrichment heatmap selected GO-KEGG analyses using Metascape. DEGs – differently expressed genes; GSEA – Gene Set Enrichment Analysis; GO – Gene Ontology; KEGG – Kyoto Encyclopedia of Genes and Genomes.
Figure 2. Gene functional enrichment analysis of the MEyellow model DEGs based on GSEA and Metascape. (A) GSEA-based GO analyses. (B) GSEA-based KEGG analyses. (C) Enrichment GO-KEGG color by cluster analyses using Metascape. (D) Enrichment GO-KEGG Color by P-value analyses using Metascape. (E) Enrichment heatmap selected GO-KEGG analyses using Metascape. DEGs – differently expressed genes; GSEA – Gene Set Enrichment Analysis; GO – Gene Ontology; KEGG – Kyoto Encyclopedia of Genes and Genomes. Figure 3. Relationship between DEGs. (A) The PPI network; the more the number of connections, the more likely the interaction. (B) Common hub genes identified using different algorithms. (C) Common hub genes of the PPI network. (D) Relationship to AD group and control group related to hub genes based on the CTD. DEGs – differently expressed genes; PPI – protein–protein interactions; AD – Alzheimer’s disease; CTD – Comparative Toxicogenomics Database.
Figure 3. Relationship between DEGs. (A) The PPI network; the more the number of connections, the more likely the interaction. (B) Common hub genes identified using different algorithms. (C) Common hub genes of the PPI network. (D) Relationship to AD group and control group related to hub genes based on the CTD. DEGs – differently expressed genes; PPI – protein–protein interactions; AD – Alzheimer’s disease; CTD – Comparative Toxicogenomics Database. Figure 4. Functional and pathway enrichment analyses of miRNAs which could regulate hub genes. (A) BP analyses. (B) CC analyses. (C) MF analyses. (D) KEGG analyses. miRNAs – microRNAS; BP – biological processes; CC – cellular components; MF – molecular functions; KEGG – Kyoto Encyclopedia of Genes and Genomes.
Figure 4. Functional and pathway enrichment analyses of miRNAs which could regulate hub genes. (A) BP analyses. (B) CC analyses. (C) MF analyses. (D) KEGG analyses. miRNAs – microRNAS; BP – biological processes; CC – cellular components; MF – molecular functions; KEGG – Kyoto Encyclopedia of Genes and Genomes. Figure 5. ROC curves of the hub genes. ROC – receiver operating characteristic.
Figure 5. ROC curves of the hub genes. ROC – receiver operating characteristic. Tables
 Table 1. Summary of Alzheimer’s disease (AD) microarray datasets from different Gene Expression Omnibus (GEO) datasets.
Table 1. Summary of Alzheimer’s disease (AD) microarray datasets from different Gene Expression Omnibus (GEO) datasets. Table 2. Gene Set Enrichment Analysis (GSEA)-based Gene Ontology (GO) snalysis.
Table 2. Gene Set Enrichment Analysis (GSEA)-based Gene Ontology (GO) snalysis. Table 3. Gene Set Enrichment Analysis (GSEA)-based and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis.
Table 3. Gene Set Enrichment Analysis (GSEA)-based and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. Table 4. Summary of hub genes.
Table 4. Summary of hub genes. Table 5. Summary of MiRNAs that regulate hub genes.
Table 5. Summary of MiRNAs that regulate hub genes. Table 6. Summary of hub gene regulation connectivity map.
Table 6. Summary of hub gene regulation connectivity map. Table 7. Hub genes and their predicted effects on Alzheimer’s disease based on univariate logistic proportional regression analysis.
Table 7. Hub genes and their predicted effects on Alzheimer’s disease based on univariate logistic proportional regression analysis. Table 1. Summary of Alzheimer’s disease (AD) microarray datasets from different Gene Expression Omnibus (GEO) datasets.
Table 1. Summary of Alzheimer’s disease (AD) microarray datasets from different Gene Expression Omnibus (GEO) datasets. Table 2. Gene Set Enrichment Analysis (GSEA)-based Gene Ontology (GO) snalysis.
Table 2. Gene Set Enrichment Analysis (GSEA)-based Gene Ontology (GO) snalysis. Table 3. Gene Set Enrichment Analysis (GSEA)-based and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis.
Table 3. Gene Set Enrichment Analysis (GSEA)-based and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. Table 4. Summary of hub genes.
Table 4. Summary of hub genes. Table 5. Summary of MiRNAs that regulate hub genes.
Table 5. Summary of MiRNAs that regulate hub genes. Table 6. Summary of hub gene regulation connectivity map.
Table 6. Summary of hub gene regulation connectivity map. Table 7. Hub genes and their predicted effects on Alzheimer’s disease based on univariate logistic proportional regression analysis.
Table 7. Hub genes and their predicted effects on Alzheimer’s disease based on univariate logistic proportional regression analysis. In Press
06 Mar 2024 : Clinical Research
Prevalence and Variation of Medical Comorbidities in Oral Surgery Patients: A Retrospective Study at Jazan ...Med Sci Monit In Press; DOI: 10.12659/MSM.943884
08 Mar 2024 : Clinical Research
Evaluation of Foot Structure in Preschool Children Based on Body MassMed Sci Monit In Press; DOI: 10.12659/MSM.943765
15 Apr 2024 : Laboratory Research
The Role of Copper-Induced M2 Macrophage Polarization in Protecting Cartilage Matrix in OsteoarthritisMed Sci Monit In Press; DOI: 10.12659/MSM.943738
07 Mar 2024 : Clinical Research
Knowledge of and Attitudes Toward Clinical Trials: A Questionnaire-Based Study of 179 Male Third- and Fourt...Med Sci Monit In Press; DOI: 10.12659/MSM.943468
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952








