09 March 2021: Lab/In Vitro Research
Effects of Pretreatment with Using 16S Ribosomal RNA Gene Sequencing in a Mouse Model of Acute Colitis Induced by Dextran Sulfate Sodium
Yi-jie Weng1BCDEF, Dan-xian Jiang2BCDEF, Jian Liang1BCDF, Shi-cai Ye1BCD, Wen-kai Tan1BCF, Cai-yuan Yu1BCF, Yu Zhou1ACDEG*DOI: 10.12659/MSM.928478
Med Sci Monit 2021; 27:e928478
Abstract
BACKGROUND: Bifidobacterium is a potentially effective and safe treatment for patients with inflammatory bowel disease (IBD), including ulcerative colitis and Crohn’s disease. However, information on the influence of B. bifidum on gut microbial diversity of treated and pretreated IBD patients is limited.
MATERIAL AND METHODS: Our study investigated therapeutic and preventive effects of B. bifidum ATCC 29521 on C57BL/6 mice with dextran sulfate sodium (DSS)-induced acute colitis via 16S ribosomal ribonucleic acid (rRNA) gene sequencing.
RESULTS: Treatment and pretreatment of mice with B. bifidum ATCC 29521 significantly alleviated the severity of acute colitis on the basis of clinical and pathologic indicators. 16S rRNA gene sequencing showed that administration of B. bifidum shifted composition of the gut microbiome in mice with DSS-induced colitis in both treated and pretreated groups. Mice pretreated with B. bifidum ATCC 29521 for 21 days exhibited a significant increase in diversity of the gut microbiome. Principal coordinate analysis showed that gut microbiota structure was shaped by different treatments and time points. On the basis of linear discriminant analysis of effect size, the abundance of the genus Escherichia-Shigella, belonging to the family Enterobacteriaceae, was reduced in the B. bifidum-treated group, indicating that pathogens were inhibited by the B. bifidum treatment. Furthermore, the genera Intestinimonas and Bacteroides were significantly associated with the B. bifidum-pretreated group.
CONCLUSIONS: 16S rRNA gene sequencing showed that pretreatment with B. bifidum ATCC 29521 reduced intestinal inflammation and altered the gut microbiota to favor the genera Intestinimonas and Bacteroides.
Keywords: Colitis, Ulcerative, Encephalocele, Inflammatory Bowel Diseases, Sequence Analysis, DNA, Bacteria, Bifidobacterium bifidum, Colitis, Colon, Dextran Sulfate, Feces, Probiotics
Background
Inflammatory bowel disease (IBD) describes a group of chronic and recurrent intestinal inflammatory diseases, including ulcerative colitis (UC) and Crohn’s disease (CD). Although the etiology remains largely unknown, a reasonable hypothesis is that the intestinal microbiota or gut microbiome activates abnormal and hyperactive immune responses in a genetically susceptible host, resulting in chronic inflammation [1,2]. Gut microbiota, consisting of many microbial species and bacteria, makes up a protective system for the gastrointestinal tract [3]. The gut microbiome, as a microbial community, mainly includes viruses, bacteria, and fungi [4]. Current treatment paradigms for IBD are based on the use of 5-aminosalicylic acid, corticosteroids, thiopurines, and infliximab, among other agents, but these standard protocols are not satisfactory; that is, a drug treatment with a nonrelapsing cure rate is not available for IBD. Therefore, nontoxic treatment choices have been increasingly used to improve IBD-associated intestinal inflammation.
Probiotics are defined as live microorganisms that provide health benefits to the host when administered in adequate amounts (Food and Agriculture Organization/World Health Organization, 2002). Different probiotic strains have been used as selective medications to alleviate the symptoms of IBD [5–8]. The mechanisms involved in the host-beneficial effects are numerous, including inhibiting the growth of pathogenic microorganisms, competing with pathogens for nutrients and niches to prevent colonization of the host by pathogens, rectifying intestinal microbial imbalance, regulating the immune system, and producing substances that contribute to cell proliferation and maturation [9–11]. Increasing the number of probiotics in the gut, such as mucosa-associated
Therefore, this study aimed to use 16S rRNA gene sequencing to investigate effects of pretreatment for 21 days with
Material and Methods
ANIMALS:
Eighty female specific-pathogen-free C57BL/6 mice (9–10 weeks old, 20±2 g) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). The mice were maintained under the same conditions, with a constant temperature (20–25°C) and humidity (40–70%). Food (chow; Teklad 7964 rodent diet, Harlan, Madison, WI), water, bedding, and cages were autoclaved.
All experiments were approved by the Guangdong Medical University Animal Experimental Ethics Committee.
BACTERIAL STRAINS AND CULTURE CONDITIONS:
INDUCTION OF EXPERIMENTAL COLITIS AND BACTERIAL COLONIZATION IN MICE:
DSS (36–50 kDa; MP Biomedicals, Aurora, OH) was dissolved in distilled water at a concentration of 3% (w/v) and was administered to the mice for 7 days to induce acute colitis. Bifidobacterium bifidum ATCC 29521 pellets were collected by centrifugation, after which the cells were washed and resuspended in saline. As shown in Figure 1, the 80 mice were randomly divided into 2 groups, a treated group and a pretreated group, with each group then being randomly divided into 2 subgroups of 20 mice each; these subgroups were treated with saline or B. bifidum ATCC 29521. The subgroups treated with saline or B. bifidum were named the treatment-with-saline (treated SL) group and treatment-with-B. bifidum ATCC 29521 (treated BB) group, respectively, whereas in the pretreated group, the respective groups were named the pretreatment-with-saline (pretreated SL) group and pretreatment-with-B. bifidum ATCC 29521 (pretreated BB) group. The mice in the treated BB group were treated with B. bifidum for 2 weeks, starting at the time of DSS administration (which lasted for 7 days), with the pretreated BB group receiving treatment for 3 weeks before DSS administration; the respective number of days of treatment thus amounted to 14 and 35 days. The mice in the test groups were also colonized with B. bifidum ATCC 29521 at a dose of 1×109 colony-forming units resuspended in 0.5 mL of saline, administered once daily by oral gavage. The mice in the control groups were given 0.5 mL of saline by oral gavage.
The feces of the mice in the pretreated group were collected at days 0, 21, and 35 (before B. bifidum colonization [preB BB] and before the saline group [preB SL]) after 3 weeks of B. bifidum colonization (preA BB) and saline (preA SL) and on the day of sacrifice (preT BB, preT SL). However, the feces of the mice in the treated group were only collected at day 14, the day of sacrifice. The mice were anesthetized with diethyl ether via intraperitoneal injection (3 mL/kg) and were euthanized at day 14 or 35 in the treated and pretreated groups, respectively (Figure 1).
WEIGHT AND CLINICAL SCORE ASSESSMENT:
The mice were weighed every day, and their clinical scores were evaluated accordingly. Weight data are shown as the percentage of the initial weight. The clinical activity score (CAS) [22] was determined on the basis of body weight, stool consistency, and occult blood. Weight loss was scored as 0, no weight loss; 1, 1–5%; 2, 5–10%; 3, 10–15%; or 4, >15%. Stool consistency was scored as 0, well-formed pellets; 2, pasty and semiformed stools that did not stick to the anus; or 4, watery diarrhea that did stick to the anus. Intestinal bleeding was scored as 0, hemoccult negative; 2, hemoccult positive; or 4, gross bleeding. The individual scores were added and divided by 3, resulting in a CAS that ranged from 0 to 4.
HISTOPATHOLOGY:
Mouse colons were fixed with 10% paraformaldehyde, embedded in paraffin using a standard protocol, cut into 4-μm sections, stained with hematoxylin and eosin, and assessed under a light microscope. Histopathologic changes were scored as follows: depth of the ulcer: 0, no ulcer; 1, mucosal involvement; 2, mucosal-submucosal involvement; 3, penetration of the muscularis propria; or 4, full-thickness involvement. Extent of the ulcer: 0, no ulcer; 1, punctate; 2, minimal; 3, moderate; or 4, widespread. Presence of inflammation: 0, none; 1, minimal; 2, mild; 3, moderate; or 4, severe. Extent of inflammation: 0, none; 1, mucosal; 2, mucosal-submucosal involvement; 3, mucosal-submucosal muscle penetration; or 4, full-thickness involvement. Location of fibrosis: 0, none; 1, mucosa only; 2, mucosa and submucosa; 3, including the muscle layer; or 4, full-thickness fibrosis. For each mouse, the scores from the 5 parts were added to obtain histologic scores that ranged from 0 to 20 [23].
DEOXYRIBONUCLEIC ACID (DNA) EXTRACTION AND 16S RRNA GENE SEQUENCING:
Total DNA was isolated from the feces using a stool DNA extraction kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The 16S rRNA gene sequencing was carried out as described by a previous study [24], with a few modifications. The 16S rRNA gene was sequenced in a region that targeted the V4 variable region, and the bacterial genome was sequenced on an Illumina HiSeq instrument with the barcoded primers 515F (GTGCCAGCMGCCGCGGTAA) and 806R (GGACTACHVGGGTWTCTA AT) according to protocols provided by Novogene Bioinformatics Technology (Beijing, China). Paired-end reads, 250 base pairs in length in each direction, were generated, and the overlapping reads were assembled.
STATISTICAL ANALYSIS:
Weight data are shown as the percentage of the initial weight. Differences in weight loss and histologic scores obtained from the groups were examined using an independent-sample t test. Single-factor repeated measures and analysis of variance were performed to evaluate CAS. After low-quality sequences were filtered out, the high-quality sequences were analyzed. The assembled reads were clustered into operational taxonomic units (OTUs) using scripts from UPARSE software (v7.0.1090), defined at a 97% similarity level. The Shannon, Chao 1, and ACE indexes were used to evaluate the α diversity, and the β diversity was assessed by principal coordinate analysis (PCoA). A comparison of the relative abundances between groups was performed by the nonparametric Mann-Whitney U test. Microbial diversity and statistical analyses were implemented in Quantitative Insights into Microbial Ecology (QIIME) and SPSS 19.0 version. P<0.05 was considered statistically significant. Differentially abundant features were determined using linear discriminate analysis effect size (LEfSe). LEfSe is an algorithm for high-dimensional biomarker discovery and explanation that identifies genomic features by characterizing the differences between 2 or more biological conditions. The threshold on the logarithmic local-density approximation score for discriminative features was 4.0.
Results
FECAL CONDITION AND BODY WEIGHT:
All mice survived throughout the whole process of the experiment. All mice showed weight loss and hematochezia, and several had diarrhea after the DSS treatment. According to the weight curve, DSS treatment led to a peak decrease in body weight on days 10 and 11, followed by a gradual return to baseline levels. After 2 weeks of B. bifidum administration, no significant difference was observed between control and B. bifidum-treated mice in the treated group. In the pretreated group, from days 31 to 34, the DSS-induced decrease in body weight was significantly less in mice treated with B. bifidum compared with the control groups (Figure 2A, 2B). Clinical symptoms were also assessed using CAS. Compared with the control groups, the mice treated with B. bifidum showed better-formed stools with less evident blood in both the treated and the pretreated groups. After exposure to DSS, the CAS curve notably increased on day 4 and then gradually decreased on day 9 after reaching its peak. After treatment with B. bifidum, the CASs for mice in both the treated and the pretreated groups were significantly decreased compared with the values for the control mice with colitis (P<0.05). Particularly, mice pretreated with B. bifidum exhibited a more significant decrease in CAS compared with the control group (P=0.000) (Figure 2C, 2D).
:
Mice with DSS-induced colitis displayed a shorter intestinal length due to intestinal inflammation. Intestinal shortening was curtailed in mice treated or pretreated with B. bifidum when compared with that in the control groups, although significant differences were not observed (treated SL group [6.9±0.6 cm] vs treated BB group [7.1±0.4 cm] and pretreated SL group [7.0±0.3 cm] vs pretreated BB group [7.3±0.5 cm]) (Figure 3A).
Pathologic inflammation was assessed using histologic colonic severity scores that combined the extent and depth of inflammation and ulceration. In both the treated and the pretreated groups, the histologic colonic severity scores were higher in mice that received saline than in those that received B. bifidum (treated SL group [9.3±2.4] vs treated BB group [7.4±2.9] and pretreated SL group [9.5±1.9] vs pretreated BB group [5.9±2.1]) (Figure 3B). In addition, the histologic scores were significantly lower in mice pretreated with B. bifidum than those in control mice (P<0.05).
Histologic inflammation in mice with DSS-induced colitis included erosion, crypt shortening or disappearance, edema, and neutrophilic infiltrates in the mucosa and lamina propria. Mice in the control groups showed increased infiltration of inflammatory cells within the epithelium (primarily neutrophils), disappearance of glandular epithelium, moderate to severe erosion of lamina propria mucosa, and even ulcerative damage. Mice treated with B. bifidum tended to show less severe effects as evidenced by lower inflammatory cell infiltration and inconsecutive erosion of epithelium (Figure 3C–3F).
OVERVIEW OF SEQUENCING ANALYSIS:
Stool samples were collected from each mouse at different time points and then pooled for the mice from the same group and same time points before DNA extraction. In total, 32 DNA samples were sequenced using a MiSeq instrument. After controlling for the quality of the raw sequences, we obtained 534 506 and 1 493 777 high-quality 16S rRNA gene sequences for the treated and the pretreated groups, respectively, with respective averages of 66 813 and 62 241 sequences per sample. Using a 97% identity threshold, the sequences were clustered into 3685 and 6176 OTUs, respectively, with respective averages of 461 and 527 OTUs per sample.
The treated and pretreated groups shared 8 predominant bacterial phyla: Bacteroidetes, Firmicutes, Proteobacteria, Deferribacteres, Tenericutes, Cyanobacteria, Verrucomicrobia, and Actinobacteria. Additional predominant bacterial phyla in the treated group included Saccharibacteria and Deinococcus-Thermus, and in the pretreated group were Chloroflexi and Synergistetes. The dominant bacterial phyla in both groups were Bacteroidetes, Firmicutes, and Proteobacteria, which accounted for over 90% of the observed phyla (Figures 4A, 5A). At the genus level, the 2 groups shared 6 predominant bacterial genera: Bacteroides, Mucispirillum, Lachnospiraceae_NK4A136_group, Helicobacter, Escherichia-Shigella, Lactobacillus, and Alistipes. Additional bacterial genera that characterized the treated group were Mycoplasma, Ruminiclostridium, and Oscillibacter, and in the pretreated group were Alloprevotella, Desulfovibrio, and Parabacteroides.
:
In the treated group, mice with acute DSS-induced colitis received B. bifidum supplements for only 2 weeks. Although no significant difference was observed in the diversity of the gut microbiota, the number of unique OTUs and the Chao 1 and Shannon indexes of mice treated with B. bifidum were higher than those observed in the control group (Figure 4B).
Of the top 10 most predominant phyla, the relative abundance of Firmicutes was more enriched in the B. bifidum-treated group than that in the control group, whereas Proteobacteria, Deferribacteres, and Tenericutes were less enriched. The abundances of Actinobacteria and Bifidobacteriaceae were also significantly higher in the B. bifidum-treated group than those in the control group, with the abundance of Actinobacteria increasing from 0.067% to 0.29% and the abundance of Bifidobacteriaceae increasing from 0.042% to 0.28%, respectively. At the genus level, the abundance of Bifidobacterium was too low to identify any differences. We performed LEfSe to determine the differential abundances of the microbial communities between the 2 groups. Mice that received saline exhibited an increased enrichment of Enterobacteriales, Enterobacteriaceae, and Escherichia-Shigella. This showed that abundances of these pathogens, which belong to phylum Proteobacteria, were significantly inhibited in mice that received B. bifidum supplements (Figure 4C, 4D).
:
Mice in the pretreated group received B. bifidum ATCC 29521 for 5 weeks to evaluate its preventative and treatment effects. The gut microbial community is considered a key indicator of the overall health of the host and is influenced by the health of the host [25]. In a comparison of 3 time points in the B. bifidum-pretreated group, 472 OTUs were observed to be shared. The most unique OTUs were identified at the time point at which the mice had been administered B. bifidum for 3 weeks, before the DSS treatment (preA BB, day 21), followed by the time point at which the mice were sacrificed (preT BB, day 35) and the baseline, at which time mice were untreated (preB BB, day 0). In a horizontal comparison, more unique OTUs were identified in the B. bifidum group than those in the saline group at the time the mice were sacrificed. In a longitudinal comparison, the gut microbiota was more diverse in mice after 3 weeks of treatment with B. bifidum than in mice at the time of sacrifice. The observed species matrix, which was simplified to the number of unique OTUs, showed an obvious increase in the preA BB group. The a diversity in the community was estimated using the Chao 1 and Shannon indexes, which were all significantly higher in mice that received B. bifidum supplements for 3 weeks than those in mice assessed on day of sacrifice (Figure 5B).
According to PCoA, the gut microbial community shifted between the B. bifidum-pretreated and saline-pretreated groups and varied among the different time points (Figure 5C). When the mice were sacrificed, mice that received the B. bifidum treatment were separated from mice that received the saline treatments. In particular, the preA BB group was distinctly separated from mice that received B. bifidum treatment at other time points. Meanwhile, the microbial communities of the untreated mice and the mice treated with 3 weeks of saline supplementation were clustered together. A structural comparison of the gut microbiota among different groups was also assessed on the basis of samples that clustered in a pair-wise similarity matrix with hierarchical clustering according to their OTUs. Notably, groups initially clustered by different time points and the distance between the preA BB and the preT BB groups was shorter than distance between preA SL and preT SL groups (Figure 5D). Therefore, the structure of the gut microbiota was shaped by different treatments and time points.
We also observed longitudinal changes in the
Furthermore, we used LEfSe to identify differentially abundant features in B. bifidum-pretreated and saline-pretreated groups at time of sacrifice. Mucispirillum, which belongs to the family Deferribacteraceae, was enriched in the saline-pretreated group at the time of sacrifice, whereas Intestinimonas and Bacteroides were enriched in the B. bifidum-pretreated group. When we compared the B. bifidum-pretreated group with the saline-pretreated group after 3 weeks of B. bifidum supplementation, the former group was enriched for Cyanobacteria, Actinobacteria, and Fusobacteria, and the latter group was enriched for Deferribacteres. Regardless of whether the mice received 3 weeks or 5 weeks of B. bifidum supplementation before or after DSS treatment, the abundance of Bifidobacterium was not significantly different between the B. bifidum-pretreated and saline-pretreated groups (Figure 5E, 5F).
Discussion
The present study determined that treatment with
Whether there is a sufficiently preventive and therapeutic approach for IBD that does not allow recurrence is currently under investigation. Conventional therapies, including 5-aminosalicylic acid, corticosteroids, and thiopurines [26], and new therapies, such as infliximab and adalimumab, have been shown to reduce inflammation and relieve symptoms, but many patients become refractory to the treatments during the course of their disease and are unable to bear the economic burden. Commensal enteric bacteria in patients with IBD are characterized by increased numbers of opportunistic pathogens, such as
In the present study, we evaluated the anti-inflammatory activity of
Most current probiotics are sold as food or nutritional supplements to improve individuals’ health. The development of probiotics is rarely based on the goal of preventing or treating a specific disease. Because of their limited purity and viability, the curative efficacy of commercial probiotics is difficult to determine [36]. Compared with other probiotics,
Celiberto et al [44] observed an effect of probiotic beverage consumption (
A few limitations should be stated to improve further investigations. First, the association between severity of acute colitis and the gut microbiome has not been analyzed. Second, the mechanism for
Conclusions
In the present mouse model of acute colitis, 16S rRNA gene sequencing showed that pretreatment with
Figures
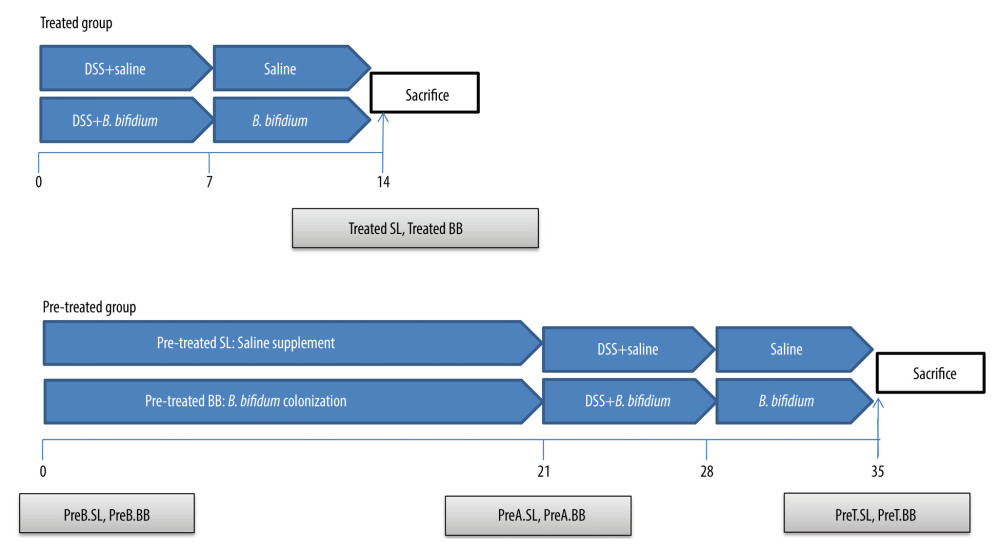 Figure 1. Experimental design for this study. (A) Mice were treated with saline or Bifidobacterium bifidum ATCC 29521 from days 0 to 14 (2 weeks) and received dextran sulfate sodium (DSS) from days 0 to 7 in the treated groups. (B) Mice were treated with saline or B. bifidum ATCC 29521 from days 21 to 35 (5 weeks) and received DSS from days 0 to 7 in the pretreated groups.
Figure 1. Experimental design for this study. (A) Mice were treated with saline or Bifidobacterium bifidum ATCC 29521 from days 0 to 14 (2 weeks) and received dextran sulfate sodium (DSS) from days 0 to 7 in the treated groups. (B) Mice were treated with saline or B. bifidum ATCC 29521 from days 21 to 35 (5 weeks) and received DSS from days 0 to 7 in the pretreated groups. 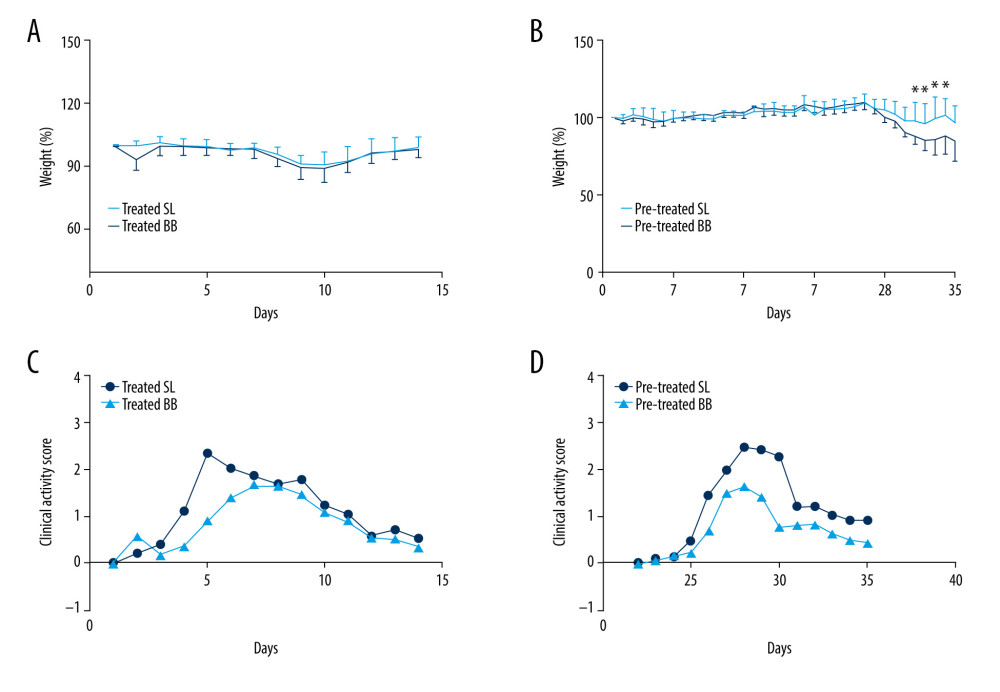 Figure 2. Weight loss and clinical activity score (CAS) of mice in the treated and pretreated groups. (A) No significant difference in weight loss was observed between the treated SL and treated BB groups. (B) Significantly less weight loss was observed in the pretreated BB group from days 31 to 34. (C) CASs of the treated BB group were remarkably reduced compared with those of the treated TL group (P=0.001). (D) CASs of the pretreated BB group were significantly decreased compared with those of the pretreated TL group (P=0.000). * P<0.05.
Figure 2. Weight loss and clinical activity score (CAS) of mice in the treated and pretreated groups. (A) No significant difference in weight loss was observed between the treated SL and treated BB groups. (B) Significantly less weight loss was observed in the pretreated BB group from days 31 to 34. (C) CASs of the treated BB group were remarkably reduced compared with those of the treated TL group (P=0.001). (D) CASs of the pretreated BB group were significantly decreased compared with those of the pretreated TL group (P=0.000). * P<0.05. 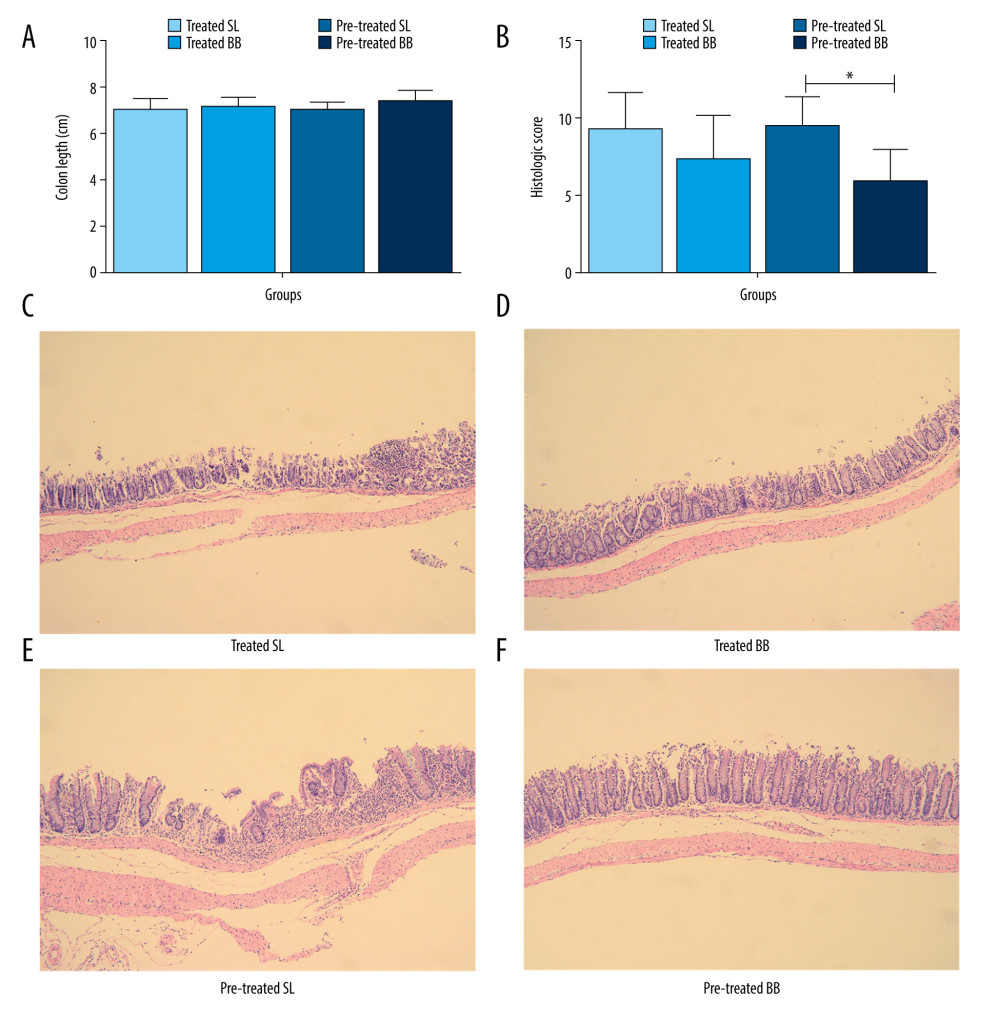 Figure 3. Colon length and histopathology analysis for effect of Bifidobacterium bifidum colonization on dextran sulfate sodium-induced colon damage. (A) Average colon length was shorter in the control groups than those in the treated groups, but the difference was not significant. (B) Histologic scores were significantly lower in the mice pretreated with B. bifidum for 5 weeks (* P<0.05). (C–F) Inflammation was more serious in the control groups than in the B. bifidum-treated and -pretreated groups. The control groups exhibited inflammatory cell infiltration, moderate to severe erosion of the lamina propria mucosa, and even ulcerative damage (hematoxylin and eosin staining, magnification ×100).
Figure 3. Colon length and histopathology analysis for effect of Bifidobacterium bifidum colonization on dextran sulfate sodium-induced colon damage. (A) Average colon length was shorter in the control groups than those in the treated groups, but the difference was not significant. (B) Histologic scores were significantly lower in the mice pretreated with B. bifidum for 5 weeks (* P<0.05). (C–F) Inflammation was more serious in the control groups than in the B. bifidum-treated and -pretreated groups. The control groups exhibited inflammatory cell infiltration, moderate to severe erosion of the lamina propria mucosa, and even ulcerative damage (hematoxylin and eosin staining, magnification ×100). 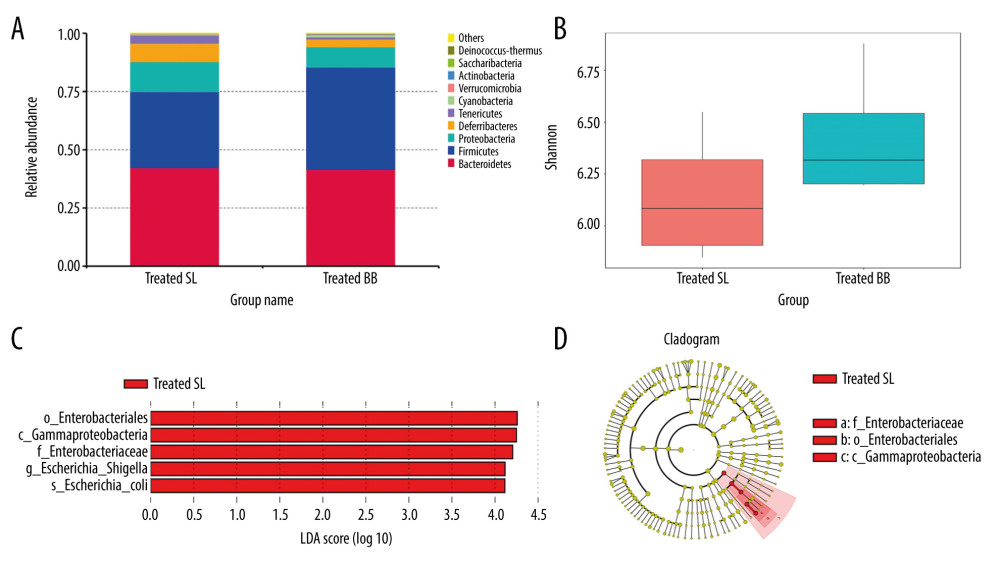 Figure 4. Changes in the microbial communities in the treated group. (A) The top 10 most frequently observed bacterial phyla in the communities. Sequences that could not be classified into any known group were labeled “other.” (B) The Shannon diversity index of the Bifidobacterium bifidum-treated group was greater than that of the control group. (C) Different features in the control and B. bifidum-treated groups were identified using linear discriminate analysis effect size, with a local-density approximation value of 4.0. (D) Differences are represented by the color of the most abundant class.
Figure 4. Changes in the microbial communities in the treated group. (A) The top 10 most frequently observed bacterial phyla in the communities. Sequences that could not be classified into any known group were labeled “other.” (B) The Shannon diversity index of the Bifidobacterium bifidum-treated group was greater than that of the control group. (C) Different features in the control and B. bifidum-treated groups were identified using linear discriminate analysis effect size, with a local-density approximation value of 4.0. (D) Differences are represented by the color of the most abundant class. 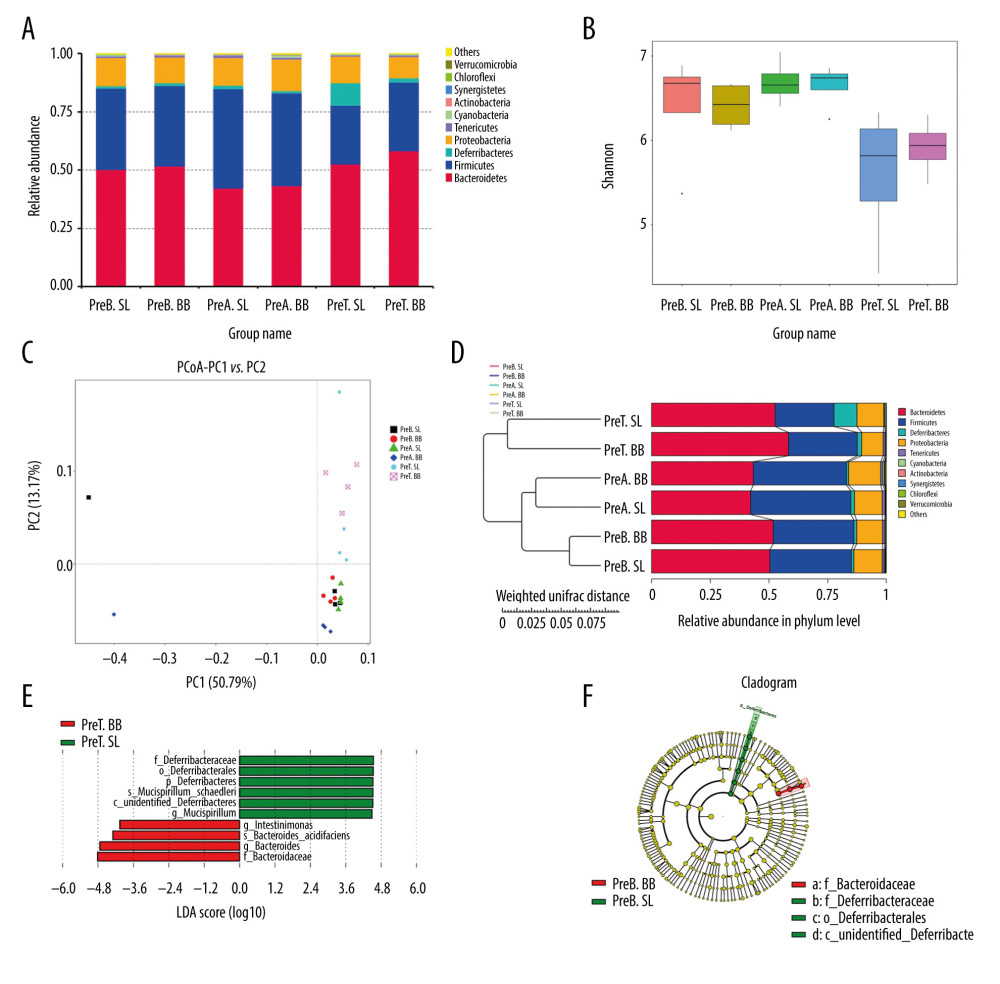 Figure 5. Changes in the microbial communities in the pretreated group. (A) The top 10 most frequently observed bacterial phyla in the communities. (B) The Shannon diversity index was higher in mice that received Bifidobacterium bifidum supplements for 3 weeks than in mice assessed on the day of sacrifice. The principal coordinate analysis (C) and unweighted pair group method with arithmetic mean tree (D) showed that the microbiota was initially clustered by different time points and treatments. Different features in the control and B. bifidum-pretreated groups on the day of sacrifice were identified using linear discriminate analysis effect size, with a local-density approximation value of 4.0 (E, F).
Figure 5. Changes in the microbial communities in the pretreated group. (A) The top 10 most frequently observed bacterial phyla in the communities. (B) The Shannon diversity index was higher in mice that received Bifidobacterium bifidum supplements for 3 weeks than in mice assessed on the day of sacrifice. The principal coordinate analysis (C) and unweighted pair group method with arithmetic mean tree (D) showed that the microbiota was initially clustered by different time points and treatments. Different features in the control and B. bifidum-pretreated groups on the day of sacrifice were identified using linear discriminate analysis effect size, with a local-density approximation value of 4.0 (E, F). References
1. Kelsen JR, Russo P, Sullivan KE, Early onset inflammatory bowel disease: Immunol Allergy Clin North Am, 2019; 39; 63-79
2. Jeong DY, Kim S, Son MJ, Induction and maintenance treatment of inflammatory bowel disease. A comprehensive review: Autoimmun Rev, 2019; 18; 439-54
3. Mangiola F, Ianiro G, Franceschi F, Gut microbiota in autism and mood disorders: World J Gastroenterol, 2016; 22; 361-68
4. Shi N, Li N, Duan X, Interaction between the gut microbiome and mucosal immune system: Mil Med Res, 2017; 4; 14
5. Xie Y, Guo Q, Li S, Anti-inflammatory properties of Bifidobacterium longum expressing human manganese superoxide dismutase using the TNBS-induced rat model of colitis: J Microbiol Biotechnol, 2017; 8; 4014
6. Satish Kumar CS, Kondal Reddy K, Boobalan G: Res Vet Sci, 2017; 110; 40-46
7. Palumbo VD, Romeo M, Marino Gammazza A, The long-term effects of probiotics in the therapy of ulcerative colitis: A clinical study: Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub, 2016; 160; 372-77
8. Guandalini S, Sansotta N, Probiotics in the treatment of inflammatory bowel disease: Adv Exp Med Biol, 2019; 1125; 101-7
9. Goncalves P, Araujo JR, Di Santo JP, A cross-talk between microbiota-derived short-chain fatty acids and the host mucosal immune system regulates intestinal homeostasis and inflammatory bowel disease: Inflamm Bowel Dis, 2018; 24; 558-72
10. Neu J, Multiomics-based strategies for taming intestinal inflammation in the neonate: Curr Opin Clin Nutr Metab Care, 2019; 22; 217-22
11. Howarth GS, Inflammatory bowel disease, a dysregulated host-microbiota interaction: Are probiotics a new therapeutic option?: J Gastroenterol Hepatol, 2008; 23; 1777-79
12. Khan I, Ullah N, Zha L, Alteration of gut microbiota in inflammatory bowel disease. Cause or consequence? IBD treatment targeting the gut microbiome: Pathogens, 2019; 8; 126
13. Sartor RB, Microbial influences in inflammatory bowel diseases: Gastroenterology, 2008; 134; 577-94
14. Aggeletopoulou I, Konstantakis C, Assimakopoulos SF, The role of the gut microbiota in the treatment of inflammatory bowel diseases: Microb Pathog, 2019; 137; 1103774
15. Veerappan GR, Betteridge J, Young PE, Probiotics for the treatment of inflammatory bowel disease: Curr Gastroenterol Rep, 2012; 14; 324-33
16. Wirtz S, Popp V, Kindermann M, Chemically induced mouse models of acute and chronic intestinal inflammation: Nat Protoc, 2017; 12; 1295-309
17. He J, Zhang F, Jian Z: Saudi J Gastroenterol, 2020; 26; 153-59
18. Zheng B, van Bergenhenegouwen J, Overbeek S, Bifidobacterium breve attenuates murine dextran sodium sulfate-induced colitis and increases regulatory T cell responses: PLoS One, 2014; 9; e95441
19. Zheng B, van Bergenhenegouwen J, van de Kant HJ, Specific probiotic dietary supplementation leads to different effects during remission and relapse in murine chronic colitis: Benef Microbes, 2016; 7; 205-13
20. Wang BG, Xu HB, Xu F: Can J Microbiol, 2016; 62; 249-62
21. Lau SK, Woo PC, Li NK, Globicatella bacteraemia identified by 16S ribosomal RNA gene sequencing: J Clin Pathol, 2016; 59; 303-7
22. Christensen B, Gibson PR, Micic D, Safety and efficacy of combination treatment with calcineurin inhibits and vedolizumab in patients with refractory inflammatory bowel disease: Clin Gastroenterol Hepatol, 2019; 17(3); 486-93
23. Rachmilewitz D, Karmeli F, Takabayashi K, Immunostimulatory DNA ameliorates experimental and spontaneous murine colitis: Gastroenterology, 2002; 122; 1428-41
24. Wang YN, Meng XC, Dong YF, Effects of microbiota in mice with acute colitis based on 16S rRNA gene sequencing: Chin Med J (Engl), 2019; 132; 1833-42
25. Mithieux G, Gut microbiota and host metabolism: What relationship: Neuroendocrinology, 2018; 106; 352-56
26. Hindryckx P, Novak G, Can we move directly from 5-ASA to a biologic agent in ulcerative colitis?: Best Pract Res Clin Gastroenterol, 2018; 32–33; 9-15
27. Speca S, Dubuquoy L, Chronic bowel inflammation and inflammatory joint disease: Pathophysiology: Joint Bone Spine, 2017; 84; 417-20
28. Yousefi B, Eslami M, Ghasemian A, Probiotics importance and their immunomodulatory properties: J Cell Physiol, 2019; 234; 8008-18
29. Kanauchi O, Andoh A, AbuBakar S, Probiotics and paraprobiotics in viral infection: Clinical application and effects on the innate and acquired immune systems: Curr Pharm Des, 2018; 24; 710-17
30. Nishida A, Inoue R, Inatomi O, Gut microbiota in the pathogenesis of inflammatory bowel disease: Clin J Gastroenterol, 2018; 11; 1-10
31. Sood A, Midha V, Makharia GK, The probiotic preparation VSL#3 induces remission in patients with mild-to-moderately active ulcerative colitis: Clin Gastroenterol Hepatol, 2009; 7; 1202-209
32. Tursi A, Brandimarte G, Papa A, Treatment of relapsing mild-to-moderate ulcerative colitis with the probiotic VSL#3 as adjunctive to a standard pharmaceutical treatment: A double-blind, randomized, placebo-controlled study: Am J Gastroenterol, 2010; 105; 2218-27
33. Miele E, Pascarella F, Giannetti E, Effect of a probiotic preparation (VSL#3) on induction and maintenance of remission in children with ulcerative colitis: Am J Gastroenterol, 2009; 104; 437-43
34. Fan H, Du J, Liu X, Effects of pentasa-combined probiotics on the microfla structure and prognosis of patients with inflammatory bowel disease: Turk J Gastroenterol, 2019; 30; 680-85
35. Gevers D, Kugathasan S, Denson LA, The treatment-naive microbiome in new-onset Crohn’s disease: Cell Host Microbe, 2014; 15; 382-92
36. Marcobal A, Underwood MA, Mills DA, Rapid determination of the bacterial composition of commercial probiotic products by terminal restriction fragment length polymorphism analysis: J Pediatr Gastroenterol Nutr, 2008; 46; 608-11
37. Strisciuglio C, Miele E, Giugliano FP, Bifidobacteria enhance antigen sampling and processing by dendritic cells in pediatric inflammatory bowel disease: Inflamm Bowel Dis, 2015; 21; 1491-98
38. Underwood MA, Intestinal dysbiosis: Novel mechanisms by which gut microbes trigger and prevent disease: Prev Med, 2014; 65; 133-37
39. Ramare F, Nicoli J, Dabard J, Trypsin-dependent production of an antibacterial substance by a human Peptostreptococcus strain in gnotobiotic rats and in vitro: Appl Environ Microbiol, 1993; 59; 2876-83
40. Akram W, Garud N, Joshi R, Role of inulin as prebiotics on inflammatory bowel disease: Drug Discov Ther, 2019; 13; 1-8
41. Lievin V, Peiffer I, Hudault S, Bifidobacterium strains from resident infant human gastrointestinal microflora exert antimicrobial activity: Gut, 2000; 47; 646-52
42. Wills ES, Jonkers DM, Savelkoul PH, Fecal microbial composition of ulcerative colitis and Crohn’s disease patients in remission and subsequent exacerbation: PLoS One, 2014; 9; e90981
43. Samanta AK, Torok VA, Percy NJ, Microbial fingerprinting detects unique bacterial communities in the faecal microbiota of rats with experimentally induced colitis: J Microbiol, 2012; 50; 218-25
44. Celiberto LS, Bedani R, Dejani NN, Effect of a probiotic beverage consumption (Enterococcus faecium CRL 183 and Bifidobacterium longum ATCC 15707) in rats with chemically induced colitis: PLoS One, 2017; 12; e0175935
45. Ekmekciu I, von Klitzing E, Fiebiger U, The probiotic compound VSL#3 modulates mucosal, peripheral, and systemic immunity following murine broad-spectrum antibiotic treatment: Front Cell Infect Microbiol, 2017; 7; 167
46. Bedani R, Pauly-Silveira ND, Roselino MN, Effect of fermented soy product on the fecal microbiota of rats fed on a beef-based animal diet: J Sci Food Agric, 2010; 90; 233-38
47. Cavallini DC, Suzuki JY, Abdalla DS, Influence of a probiotic soy product on fecal microbiota and its association with cardiovascular risk factors in an animal model: Lipids Health Dis, 2011; 10; 126
48. Mirsepasi-Lauridsen HC, Vallance BA, Krogfelt KA, Escherichia coli pathobionts associated with inflammatory bowel disease: Clin Microbiol Rev, 2019; 32; e00060-18
49. Papa E, Docktor M, Smillie C, Non-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease: PLoS One, 2012; 7; e39242
50. Goyal A, Yeh A, Bush BR, Safety, clinical response, and microbiome findings following fecal microbiota transplant in children with inflammatory bowel disease: Inflamm Bowel Dis, 2018; 24; 410-12
51. Litvak Y, Sharon S, Hyams M, Epithelial cells detect functional type III secretion system of enteropathogenic Escherichia coli through a novel NF-κB signaling pathway: PLoS Pathog, 2017; 13; e10006472
52. Rahman K, Sasaki M, Nusrat A, Crohn’s disease-associated Escherichia coli survive in macrophages by suppressing NFkappaB signaling: Inflamm Bowel Dis, 2014; 20; 1419-25
53. Nguyen HT, Dalmasso G, Muller S, Crohn’s disease-associated adherent invasive Escherichia coli modulate levels of microRNAs in intestinal epithelial cells to reduce autophagy: Gastroenterology, 2014; 146; 508-19
54. Munyaka PM, Sepehri S, Ghia JE, Carrageenan gum and adherent invasive Escherichia coli in a piglet model of inflammatory bowel disease: Impact on intestinal mucosa-associated microbiota: Front Microbiol, 2016; 7; 462
55. Berry D, Schwab C, Milinovich G, Phylotype-level 16S rRNA analysis reveals new bacterial indicators of health state in acute murine colitis: ISME J, 2012; 6; 2091-106
56. Herp S, Brugiroux S, Garzetti D, Mucispirillum schaedleri antagonizes Salmonella virulence to protect mice against colitis: Cell Host Microbe, 2019; 25; 681-94
57. Frank DN, St Amand AL, Feldman RA, Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases: Proc Natl Acad Sci USA, 2007; 104; 13780-85
58. Park JS, Choi JW, Jhun J, Lactobacillus acidophilus improves intestinal inflammation in an acute colitis mouse model by regulation of Th17 and Treg cell balance and fibrosis development: J Med Food, 2018; 21; 215-24
59. Round JL, Mazmanian SK, Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota: Proc Natl Acad Sci USA, 2010; 107; 12204-9
60. Mazmanian SK, Round JL, Kasper DL, A microbial symbiosis factor prevents intestinal inflammatory disease: Nature, 2008; 453; 620-25
61. Din AU, Hassan A, Zhu Y: J Nutr Biochem, 2020; 79; 108353
Figures
 Figure 1. Experimental design for this study. (A) Mice were treated with saline or Bifidobacterium bifidum ATCC 29521 from days 0 to 14 (2 weeks) and received dextran sulfate sodium (DSS) from days 0 to 7 in the treated groups. (B) Mice were treated with saline or B. bifidum ATCC 29521 from days 21 to 35 (5 weeks) and received DSS from days 0 to 7 in the pretreated groups.
Figure 1. Experimental design for this study. (A) Mice were treated with saline or Bifidobacterium bifidum ATCC 29521 from days 0 to 14 (2 weeks) and received dextran sulfate sodium (DSS) from days 0 to 7 in the treated groups. (B) Mice were treated with saline or B. bifidum ATCC 29521 from days 21 to 35 (5 weeks) and received DSS from days 0 to 7 in the pretreated groups. Figure 2. Weight loss and clinical activity score (CAS) of mice in the treated and pretreated groups. (A) No significant difference in weight loss was observed between the treated SL and treated BB groups. (B) Significantly less weight loss was observed in the pretreated BB group from days 31 to 34. (C) CASs of the treated BB group were remarkably reduced compared with those of the treated TL group (P=0.001). (D) CASs of the pretreated BB group were significantly decreased compared with those of the pretreated TL group (P=0.000). * P<0.05.
Figure 2. Weight loss and clinical activity score (CAS) of mice in the treated and pretreated groups. (A) No significant difference in weight loss was observed between the treated SL and treated BB groups. (B) Significantly less weight loss was observed in the pretreated BB group from days 31 to 34. (C) CASs of the treated BB group were remarkably reduced compared with those of the treated TL group (P=0.001). (D) CASs of the pretreated BB group were significantly decreased compared with those of the pretreated TL group (P=0.000). * P<0.05. Figure 3. Colon length and histopathology analysis for effect of Bifidobacterium bifidum colonization on dextran sulfate sodium-induced colon damage. (A) Average colon length was shorter in the control groups than those in the treated groups, but the difference was not significant. (B) Histologic scores were significantly lower in the mice pretreated with B. bifidum for 5 weeks (* P<0.05). (C–F) Inflammation was more serious in the control groups than in the B. bifidum-treated and -pretreated groups. The control groups exhibited inflammatory cell infiltration, moderate to severe erosion of the lamina propria mucosa, and even ulcerative damage (hematoxylin and eosin staining, magnification ×100).
Figure 3. Colon length and histopathology analysis for effect of Bifidobacterium bifidum colonization on dextran sulfate sodium-induced colon damage. (A) Average colon length was shorter in the control groups than those in the treated groups, but the difference was not significant. (B) Histologic scores were significantly lower in the mice pretreated with B. bifidum for 5 weeks (* P<0.05). (C–F) Inflammation was more serious in the control groups than in the B. bifidum-treated and -pretreated groups. The control groups exhibited inflammatory cell infiltration, moderate to severe erosion of the lamina propria mucosa, and even ulcerative damage (hematoxylin and eosin staining, magnification ×100). Figure 4. Changes in the microbial communities in the treated group. (A) The top 10 most frequently observed bacterial phyla in the communities. Sequences that could not be classified into any known group were labeled “other.” (B) The Shannon diversity index of the Bifidobacterium bifidum-treated group was greater than that of the control group. (C) Different features in the control and B. bifidum-treated groups were identified using linear discriminate analysis effect size, with a local-density approximation value of 4.0. (D) Differences are represented by the color of the most abundant class.
Figure 4. Changes in the microbial communities in the treated group. (A) The top 10 most frequently observed bacterial phyla in the communities. Sequences that could not be classified into any known group were labeled “other.” (B) The Shannon diversity index of the Bifidobacterium bifidum-treated group was greater than that of the control group. (C) Different features in the control and B. bifidum-treated groups were identified using linear discriminate analysis effect size, with a local-density approximation value of 4.0. (D) Differences are represented by the color of the most abundant class. Figure 5. Changes in the microbial communities in the pretreated group. (A) The top 10 most frequently observed bacterial phyla in the communities. (B) The Shannon diversity index was higher in mice that received Bifidobacterium bifidum supplements for 3 weeks than in mice assessed on the day of sacrifice. The principal coordinate analysis (C) and unweighted pair group method with arithmetic mean tree (D) showed that the microbiota was initially clustered by different time points and treatments. Different features in the control and B. bifidum-pretreated groups on the day of sacrifice were identified using linear discriminate analysis effect size, with a local-density approximation value of 4.0 (E, F).
Figure 5. Changes in the microbial communities in the pretreated group. (A) The top 10 most frequently observed bacterial phyla in the communities. (B) The Shannon diversity index was higher in mice that received Bifidobacterium bifidum supplements for 3 weeks than in mice assessed on the day of sacrifice. The principal coordinate analysis (C) and unweighted pair group method with arithmetic mean tree (D) showed that the microbiota was initially clustered by different time points and treatments. Different features in the control and B. bifidum-pretreated groups on the day of sacrifice were identified using linear discriminate analysis effect size, with a local-density approximation value of 4.0 (E, F). In Press
06 Mar 2024 : Clinical Research
Comparison of Outcomes between Single-Level and Double-Level Corpectomy in Thoracolumbar Reconstruction: A ...Med Sci Monit In Press; DOI: 10.12659/MSM.943797
21 Mar 2024 : Meta-Analysis
Economic Evaluation of COVID-19 Screening Tests and Surveillance Strategies in Low-Income, Middle-Income, a...Med Sci Monit In Press; DOI: 10.12659/MSM.943863
10 Apr 2024 : Clinical Research
Predicting Acute Cardiovascular Complications in COVID-19: Insights from a Specialized Cardiac Referral Dep...Med Sci Monit In Press; DOI: 10.12659/MSM.942612
06 Mar 2024 : Clinical Research
Enhanced Surgical Outcomes of Popliteal Cyst Excision: A Retrospective Study Comparing Arthroscopic Debride...Med Sci Monit In Press; DOI: 10.12659/MSM.941102
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952








