23 January 2021: Database Analysis
Correlation Between High Expression of and Improved Overall Survival in Ovarian Cancer Patients
Hui Shang1FG, Lingyun Shi2BC, Xuena Jiang3CF, Peng Zhou4CD, Yongqing Wei1AB*DOI: 10.12659/MSM.928763
Med Sci Monit 2021; 27:e928763
Abstract
BACKGROUND: The aim of the present work was to evaluate FOXA2 expression in ovarian cancer and to use integrated bioinformatics analysis to correlate it with patient prognosis.
MATERIAL AND METHODS: FOXA2 expression was evaluated in multiple cancers in The Cancer Genome Atlas database. A protein–protein interaction (PPI) network relevant to FOXA2 was constructed using the Search Tool for Retrieval of Interacting Genes/Proteins (STRIN). Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses were performed of FOXA2 and relevant genes. Correlations between overall survival (OS), disease-free survival, and FOXA2 expression were evaluated. An immunohistochemical assay (IHC) was used to test for FOXA2 protein expression in 79 ovarian cancer specimens.
RESULTS: FOXA2 mRNA was upregulated in colorectal, stomach, liver, and endometrial cancers. In the PPI network, 21 protein nodes and 533 edges were constructed with a local clustering coefficient of 0.698, which indicated significant PPI enrichment (P<0.01). FOXA2 and relevant genes were mainly enriched in the signaling pathways regulating pluripotency of stem cells, cancer, and AMPK. A survival analysis indicated that OS was significantly longer in patients with higher versus lower FOXA2 protein expression (HR=0.73, P<0.01). The IHC assay showed that the FOXA2 protein was mainly positively expressed in the nucleoplasm of tumor cells with brown-yellow staining. Of the 79 ovarian cancer samples, 31 (39.2%) highly expressed FOXA2. The FOXA2 gene was correlated with International Federation of Gynecology and Obstetrics staging and with lymph node metastasis (both P<0.05).
CONCLUSIONS: Upregulation of the FOXA2 gene was correlated with improved OS in patients with ovarian cancer and it can be used as a prognostic biomarker and potential treatment target.
Keywords: Diagnosis, Ovarian Neoplasms, Cluster Analysis, Databases, Factual, Gene Expression Profiling, Hepatocyte Nuclear Factor 3-beta, Protein Interaction Maps
Background
Ovarian cancer is a commonly diagnosed carcinoma of the female reproductive system. Estimates indicate that in 2019, 1390 patients with ovarian cancer in the United States died of the disease [1]. Ovarian cancer is the most common cause of cancer-related deaths in women, ranking fourth in cancer-associated mortality among women in developed countries. Cancer epidemiology data from China show that the mortality rate in patients with ovarian cancer increased by 21.6% and 1.7% from 2000 to 2003 and 2003 to 2011, respectively, ranking the disease first among carcinomas in terms of deadliness [2]. The development of ovarian cancer is occult and about 75% of cases are diagnosed when the disease is advanced (International Federation of Gynecology and Obstetrics [FIGO] stages III and IV), resulting in a poor prognosis for these patients and an extremely low 5-year survival rate [3]. Therefore, it is of great importance to identify effective biological markers that can facilitate early diagnosis of ovarian cancer and improve patient prognosis [4].
FOX proteins are characterized by a conserved DNA domain that is 110 amino acids long and similar in structure and appearance to the forkhead box [5]. Both fungi and animals have been found to contain a large number of FOX family proteins, which play an important role in embryo development, cell differentiation, energy metabolism, and immune regulation [6]. The FOX family consists of 19 subfamilies, named after FOXA and FOXS, and consists of more than 100 members [7]. The most widely used and studied of them is the FOXA subfamily, which is composed of 3 members:
Material and Methods
:
PROTEIN–PROTEIN INTERACTION NETWORK CONSTRUCTION:
A protein–protein interaction (PPI) network related to FOXA2 was constructed using the Search Tool for Retrieval of Interacting Genes/Proteins (STRIN) database (http://string-db.org/cgi/input.pl) [13]. The conditions for the PPI network were: (1) confidence >0.7; and (2) source of interaction limited to co-expression, gene function, and neighborhood relationship.
:
Seventy-nine patients with ovarian cancer who underwent surgery in our hospital were included in the present study. Written informed consent was obtained from all of the individuals and the research was approved by the Ethics Committee of our hospital. Tumor tissue and corresponding normal ovarian tissue were collected intraoperatively, immediately quick frozen in liquid nitrogen, and then transferred to and stored in a freezer at −80°C until the next use. The mean age of the 79 patients was 52.6±11.3 years. The FIGO stage of ovarian cancer was I/II in 32 cases and II/IV in 47 cases. The pathologic type was mucinous, serous, and endometrioid carcinoma in 52, 21, and 6 cases, respectively.
SURVIVAL ANALYSIS:
Patients in the TCGA databases with ovarian cancer were divided into 2 groups, based on the relative level of expression of
STATISTICAL ANALYSIS:
STATA statistical software, version 11.0, was used for data evaluation. Data were expressed as numbers (n) and percentages (%) and compared with a chi-square or Fisher’s exact test. Survival data were expressed as medians and compared using a log-rank test. The correlation between
Results
:
FOXA2 mRNA expression was quite different among the cancers (Figure 1A). FOXA2 mRNA was upregulated in colorectal, stomach, liver, and endometrial cancers. The highest rate of FOXA2 protein expression was in prostate, breast, and urothelial cancers (Figure 1B). No statistically significant differences were seen in FOXA2 mRNA expression in patients whose ovarian cancer had different types of mutations (Figure 2).
PPI NETWORK ANALYSIS:
A PPI network for FOXA2 and proteins related to it was constructed using the Search Tool for Retrieval of Interacting Genes/Proteins (STRIN) database. Twenty-one protein nodes and 533 edges were identified that had a local clustering coefficient of 0.698, which indicated significant PPI enrichment (P<0.01) (Figure 3).
GENE ONTOLOGY ENRICHMENT:
In terms of biological processes, FOXA2 and genes related to it were mainly found to be enriched in development of the endocrine system, positive regulation of cellular biosynthetic processes, and development of the pancreas. Enrichment in cellular components occurred largely in the nucleus, intracellular membrane-bounded organelle, and transcription factor complex. Regarding molecular function, FOXA2 and genes related to it were concentrated in transcription regulatory-region sequence-specific DNA binding, proximal promoter sequence-specific DNA binding, and RNA polymerase II regulatory-region sequence-specific DNA binding (Table 1).
KYOTO ENCYCLOPEDIA OF GENES AND GENOMES PATHWAY ENRICHMENT:
FOXA2 and genes related to it were mainly enriched in the signaling pathways regulating pluripotency of stem cells, pathways in cancer, and the AMP-activated protein kinase (AMPK) signaling pathway (Table 2, Figure 4).
SURVIVAL ANALYSIS:
Log-rank analysis indicated that OS was significantly longer in patients with high FOXA2 expression than in those with low expression (HR=0.73, P<0.01). However, DFS (HR=0.82, P>0.05) was not statistically different between the groups with high and low expression of FOXA2 (Figure 5). In the Kaplan-Meier plotter database, we also found that OS in patients with high FOXA2 expression was significantly longer than in those with low expression (HR=0.69, P<0.01) (Figure 5).
:
An IHC assay showed that the FOXA2 protein was mainly positively expressed in the nucleoplasm of tumor cells that stained brown-yellow (Figure 6). Of the 79 ovarian cancer samples that were included, 31 (39.2%) showed increased expression of the FOXA2 protein. The FOXA2 protein was correlated with FIGO stage and lymph node metastasis (both P<0.05) (Table 3). Multivariate logistic regression of FOXA2 protein expression and clinical features of ovarian cancer indicated that increased expression of the protein was an independent factor for lymph node metastasis in patients with ovarian cancer (OR=0.36, P<0.05) (Figure 7).
Discussion
The
In the present study, we found that
The molecular mechanism through which
Conclusions
In the present study,
Figures
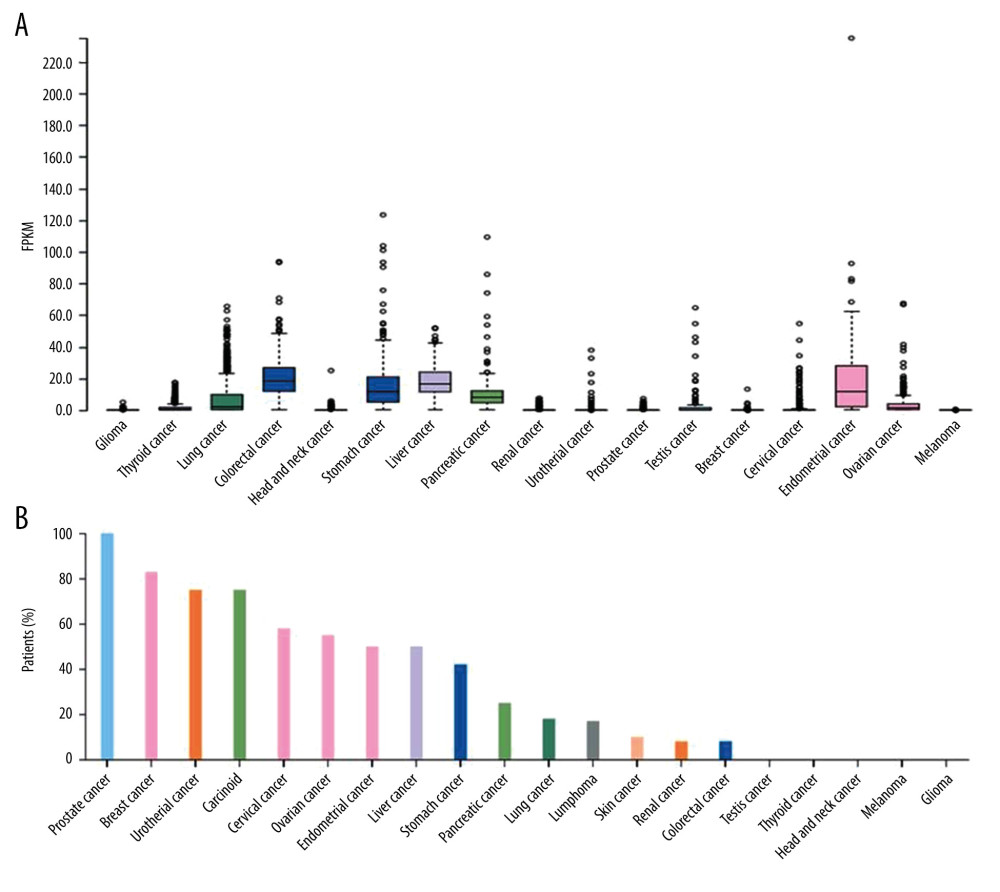 Figure 1. Bar and box plot of FOXA2 mRNA expression in tissue from multiple types of cancer. (A) Box plot expression in multiple cancer tissues. (B) Bar plot of FOXA2 protein positive expression rates in multiple cancers.
Figure 1. Bar and box plot of FOXA2 mRNA expression in tissue from multiple types of cancer. (A) Box plot expression in multiple cancer tissues. (B) Bar plot of FOXA2 protein positive expression rates in multiple cancers. 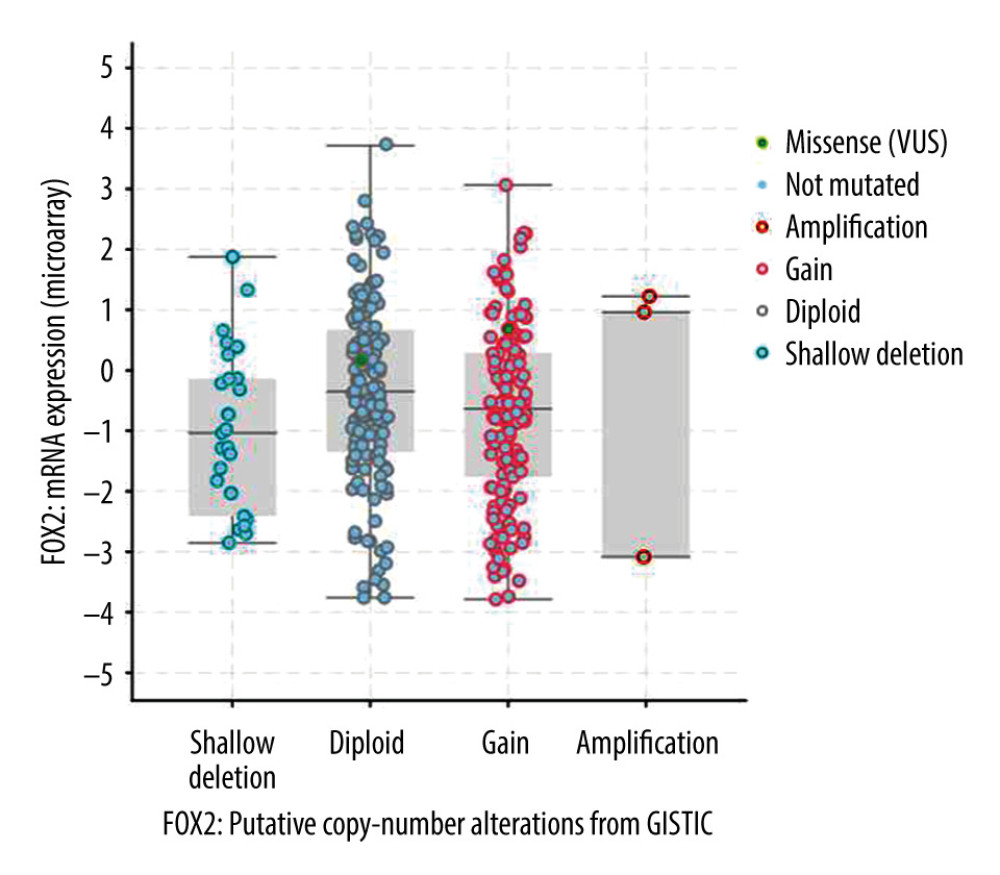 Figure 2. Scatter plot of FOXA2 mRNA expression in different types of mutations.
Figure 2. Scatter plot of FOXA2 mRNA expression in different types of mutations. 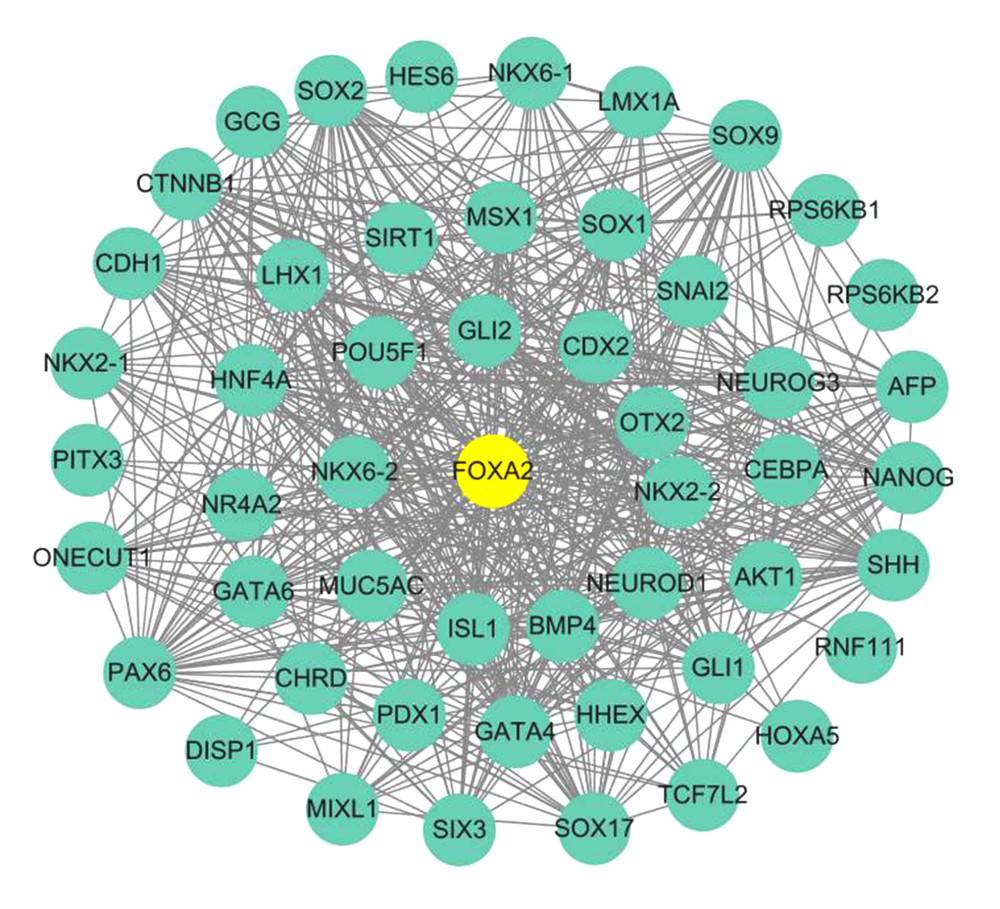 Figure 3. Protein–protein interaction network (PPI) for FOXA2 and correlated proteins.
Figure 3. Protein–protein interaction network (PPI) for FOXA2 and correlated proteins. 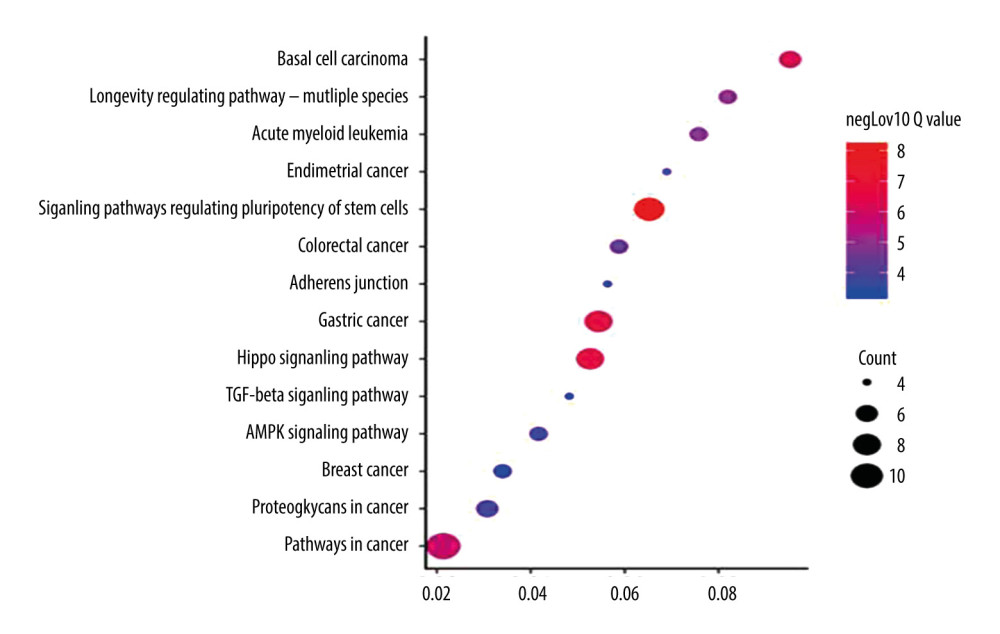 Figure 4. Bubble plot of Kyoto Encyclopedia of Genes and Genomes enrichment of FOXA2 and relevant genes.
Figure 4. Bubble plot of Kyoto Encyclopedia of Genes and Genomes enrichment of FOXA2 and relevant genes. 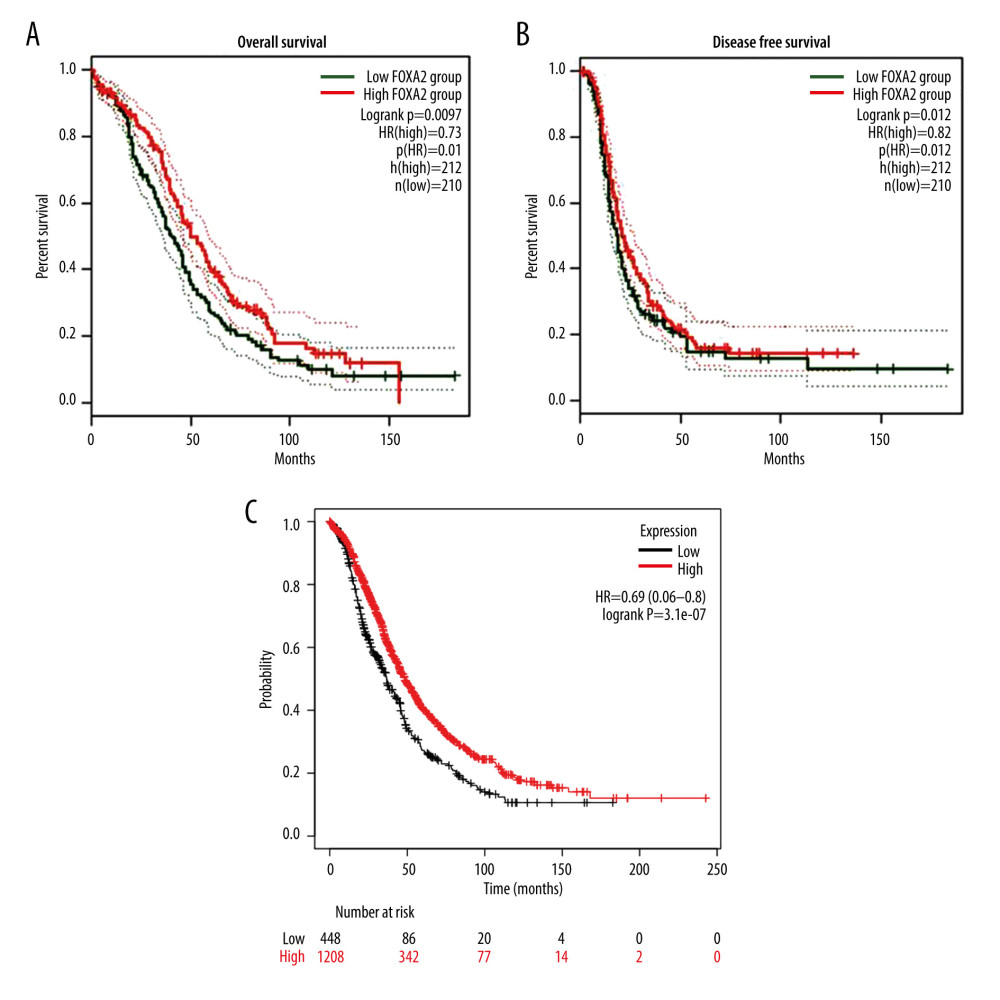 Figure 5. Overall survival (OS) and disease-free survival curve for patients with ovarian cancer who had high and low expression of FOXA2. (A) Overall survival. (B) Disease-free survival. (C) OS curve created using data in the Kaplan-Meier plotter database from patients with ovarian cancer who had high and low expression of FOXA2.
Figure 5. Overall survival (OS) and disease-free survival curve for patients with ovarian cancer who had high and low expression of FOXA2. (A) Overall survival. (B) Disease-free survival. (C) OS curve created using data in the Kaplan-Meier plotter database from patients with ovarian cancer who had high and low expression of FOXA2. 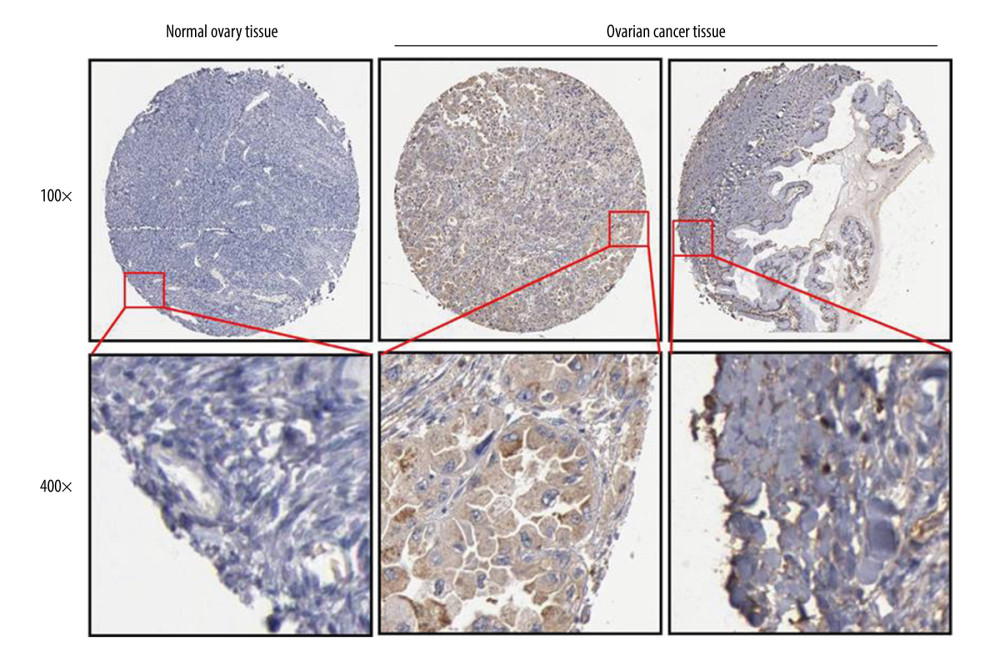 Figure 6. FOXA2 protein expression detected with an immunohistochemistry assay in normal and cancerous ovarian tissue.
Figure 6. FOXA2 protein expression detected with an immunohistochemistry assay in normal and cancerous ovarian tissue. 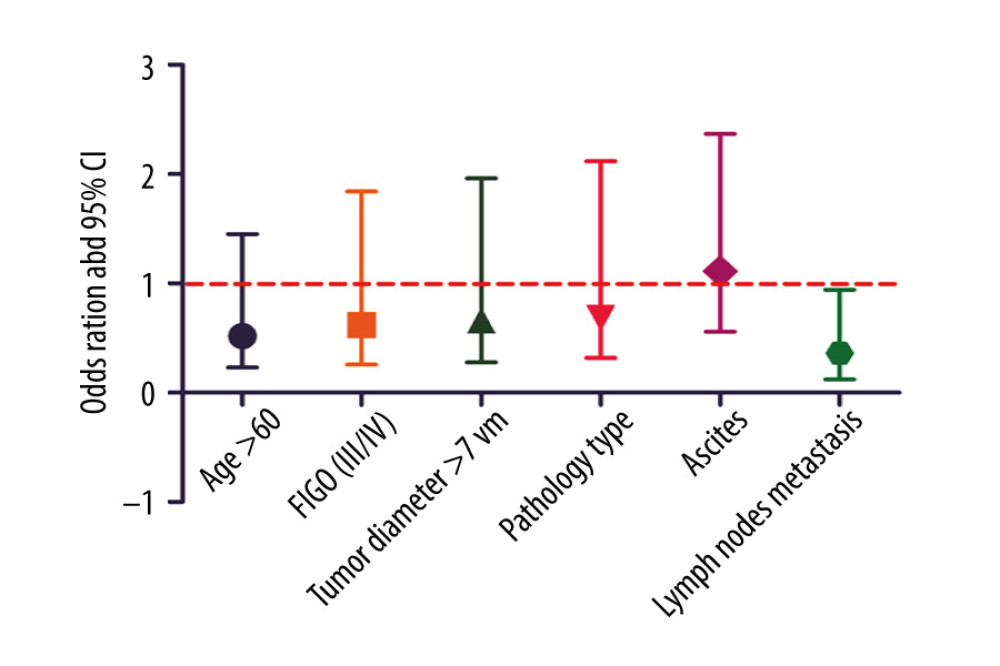 Figure 7. Logistic regression analysis of FOXA2 protein expression and clinical features in patients with ovarian cancer showed that high expression of FOXA2 protein was independently associated with lymph node metastasis.
Figure 7. Logistic regression analysis of FOXA2 protein expression and clinical features in patients with ovarian cancer showed that high expression of FOXA2 protein was independently associated with lymph node metastasis. References
1. Siegel RL, Miller KD, Jemal A, Cancer statistics, 2019: Cancer J Clin, 2019; 69; 7-34
2. Chen W, Zheng R, Baade PD, Cancer statistics in China, 2015: Cancer J Clin, 2016; 66; 115-32
3. DeSantis CE, Miller KD, Goding Sauer A, Cancer statistics for African Americans, 2019: Cancer J Clin, 2019; 69; 211-33
4. Slatnik CL, Duff E, Ovarian cancer: Ensuring early diagnosis: Nurse Pract, 2015; 40; 47-54
5. Wang J, Li W, Zhao Y, Members of FOX family could be drug targets of cancers: Pharmacol Ther, 2018; 181; 183-96
6. Williams CB, Chatila TA, Fox family ties: Cell Res, 2013; 23; 452-54
7. Katoh M, Katoh M, Human FOX gene family (review): Int J Oncol, 2004; 25; 1495-500
8. Katoh M, Igarashi M, Fukuda H, Cancer genetics and genomics of human FOX family genes: Cancer Lett, 2013; 328; 198-206
9. Kim KK, Adelstein RS, Kawamoto S, Identification of neuronal nuclei (NeuN) as Fox-3, a new member of the Fox-1 gene family of splicing factors: J Biol Chem, 2009; 284; 31052-61
10. Wang B, Liu G, Ding L, FOXA2 promotes the proliferation, migration and invasion, and epithelial mesenchymal transition in colon cancer: Exp Ther Med, 2018; 16; 133-40
11. Basseres DS, D’Alò F, Yeap BY, Frequent downregulation of the transcription factor Foxa2 in lung cancer through epigenetic silencing: Lung Cancer, 2012; 77; 31-37
12. Zhang Z, Yang C, Gao W, FOXA2 attenuates the epithelial to mesenchymal transition by regulating the transcription of E-cadherin and ZEB2 in human breast cancer: Cancer Lett, 2015; 361; 240-50
13. Szklarczyk D, Gable AL, Lyon D, STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets: Nucleic Acids Res, 2019; 47; D607-13
14. Li J, Dantas Machado AC, Guo M, Structure of the Forkhead domain of FOXA2 bound to a complete DNA consensus site: Biochemistry, 2017; 56; 3745-53
15. Burtscher I, Lickert H, Foxa2 regulates polarity and epithelialization in the endoderm germ layer of the mouse embryo: Development, 2009; 136; 1029-38
16. Popovic J, Klajn A, Petrovic I, Tissue-specific Forkhead protein FOXA2 up-regulates SOX14 gene expression: Biochim Biophys Acta, 2010; 1799; 411-18
17. Camolotto SA, Pattabiraman S, Mosbruger TL, FoxA1 and FoxA2 drive gastric differentiation and suppress squamous identity in NKX2-1-negative lung cancer: Elife, 2018; 7; e38579
18. Jang SM, An JH, Kim CH, Transcription factor FOXA2-centered transcriptional regulation network in non-small cell lung cancer: Biochem Biophys Res Commun, 2015; 463; 961-67
19. Li C, Lu S, Shi Y, MicroRNA-187 promotes growth and metastasis of gastric cancer by inhibiting FOXA2: Oncol Rep, 2017; 37; 1747-55
20. Chand V, Pandey A, Kopanja D, Opposing roles of the Forkhead box factors FoxM1 and FoxA2 in liver cancer: Mol Cancer Res, 2019; 17; 1063-74
21. Ren XY, Yang WB, Li XMNKX2-1 and FOXA2 expression in ovarian cancer and its clinical significance: Maternal Child Health Care China, 2019; 34(12); 2857-60 [in Chinese]
Figures
 Figure 1. Bar and box plot of FOXA2 mRNA expression in tissue from multiple types of cancer. (A) Box plot expression in multiple cancer tissues. (B) Bar plot of FOXA2 protein positive expression rates in multiple cancers.
Figure 1. Bar and box plot of FOXA2 mRNA expression in tissue from multiple types of cancer. (A) Box plot expression in multiple cancer tissues. (B) Bar plot of FOXA2 protein positive expression rates in multiple cancers. Figure 2. Scatter plot of FOXA2 mRNA expression in different types of mutations.
Figure 2. Scatter plot of FOXA2 mRNA expression in different types of mutations. Figure 3. Protein–protein interaction network (PPI) for FOXA2 and correlated proteins.
Figure 3. Protein–protein interaction network (PPI) for FOXA2 and correlated proteins. Figure 4. Bubble plot of Kyoto Encyclopedia of Genes and Genomes enrichment of FOXA2 and relevant genes.
Figure 4. Bubble plot of Kyoto Encyclopedia of Genes and Genomes enrichment of FOXA2 and relevant genes. Figure 5. Overall survival (OS) and disease-free survival curve for patients with ovarian cancer who had high and low expression of FOXA2. (A) Overall survival. (B) Disease-free survival. (C) OS curve created using data in the Kaplan-Meier plotter database from patients with ovarian cancer who had high and low expression of FOXA2.
Figure 5. Overall survival (OS) and disease-free survival curve for patients with ovarian cancer who had high and low expression of FOXA2. (A) Overall survival. (B) Disease-free survival. (C) OS curve created using data in the Kaplan-Meier plotter database from patients with ovarian cancer who had high and low expression of FOXA2. Figure 6. FOXA2 protein expression detected with an immunohistochemistry assay in normal and cancerous ovarian tissue.
Figure 6. FOXA2 protein expression detected with an immunohistochemistry assay in normal and cancerous ovarian tissue. Figure 7. Logistic regression analysis of FOXA2 protein expression and clinical features in patients with ovarian cancer showed that high expression of FOXA2 protein was independently associated with lymph node metastasis.
Figure 7. Logistic regression analysis of FOXA2 protein expression and clinical features in patients with ovarian cancer showed that high expression of FOXA2 protein was independently associated with lymph node metastasis. Tables
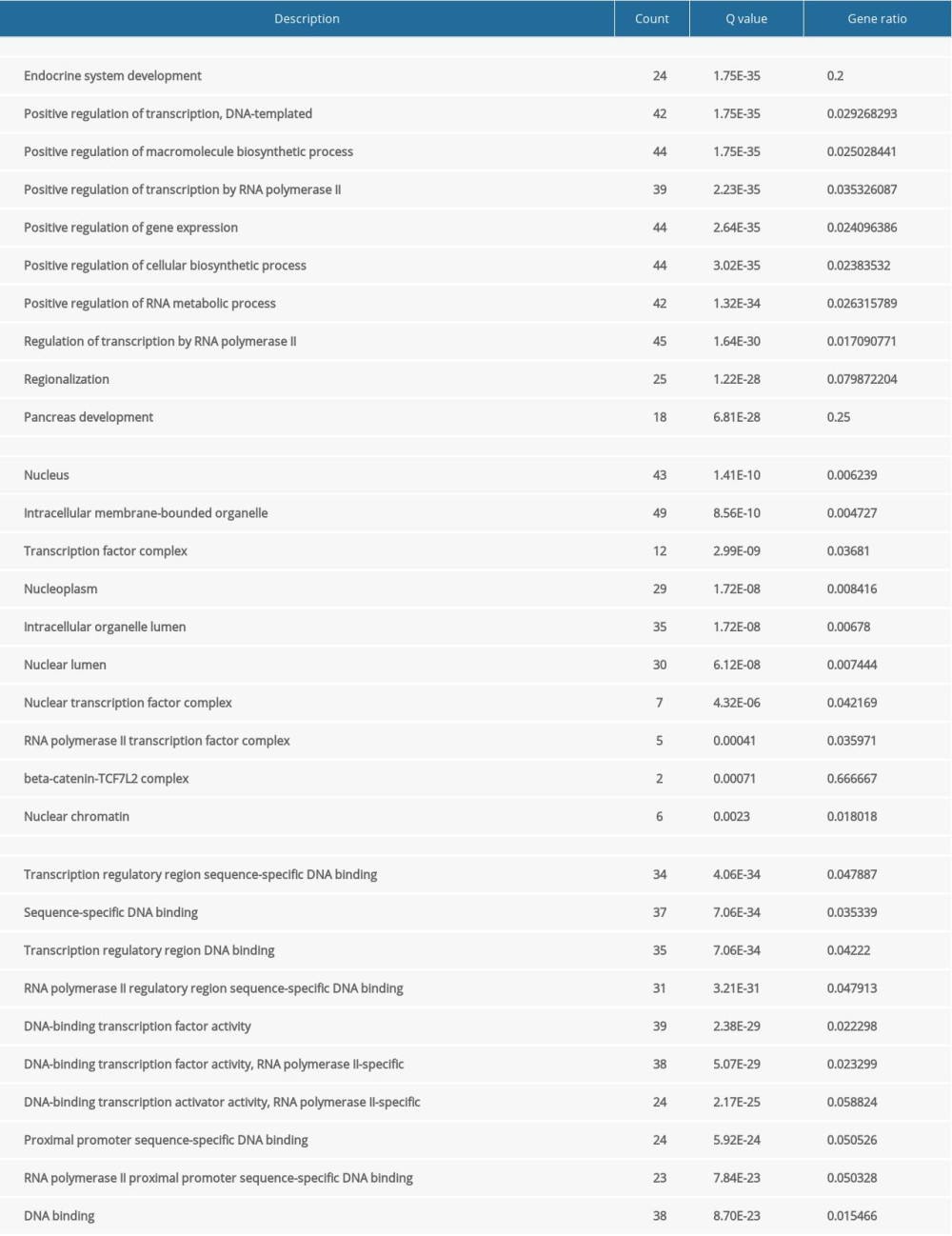 Table 1. Gene Ontology (GO) enrichment of FOXA2 and relevant genes.
Table 1. Gene Ontology (GO) enrichment of FOXA2 and relevant genes.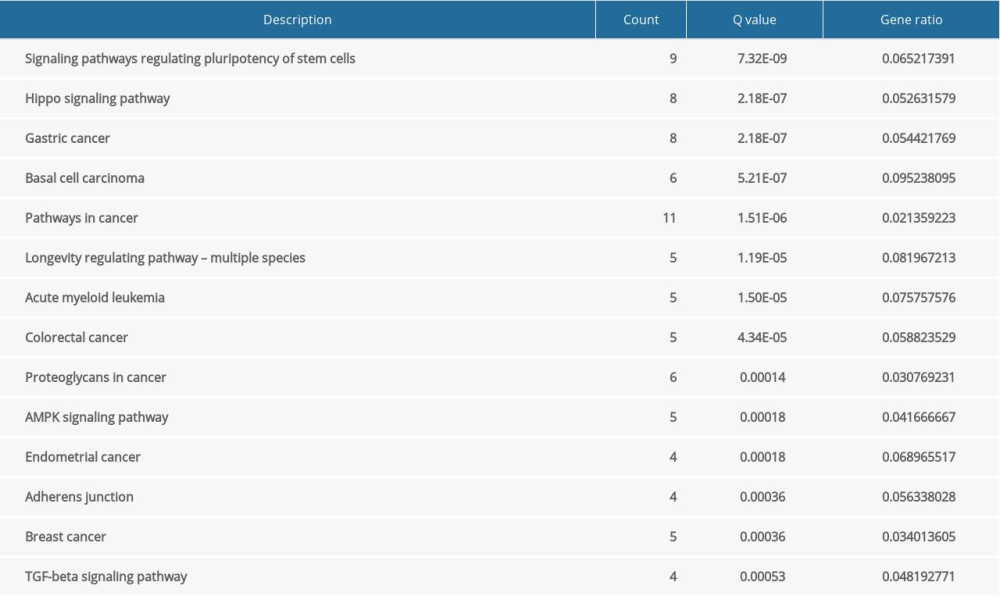 Table 2. KEGG enrichment of FOXA2 and relevant genes.
Table 2. KEGG enrichment of FOXA2 and relevant genes.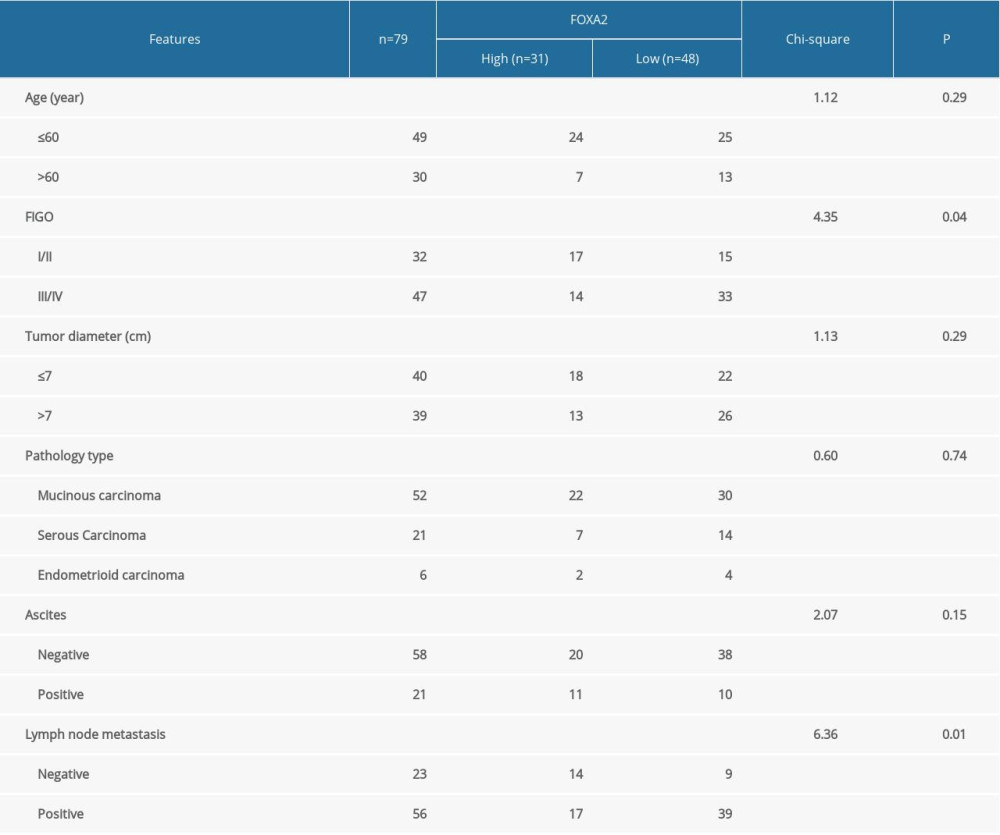 Table 3. Correlation between FOXA2 protein expression and ovarian cancer patients clinical features.
Table 3. Correlation between FOXA2 protein expression and ovarian cancer patients clinical features. Table 1. Gene Ontology (GO) enrichment of FOXA2 and relevant genes.
Table 1. Gene Ontology (GO) enrichment of FOXA2 and relevant genes. Table 2. KEGG enrichment of FOXA2 and relevant genes.
Table 2. KEGG enrichment of FOXA2 and relevant genes. Table 3. Correlation between FOXA2 protein expression and ovarian cancer patients clinical features.
Table 3. Correlation between FOXA2 protein expression and ovarian cancer patients clinical features. In Press
07 Mar 2024 : Clinical Research
Knowledge of and Attitudes Toward Clinical Trials: A Questionnaire-Based Study of 179 Male Third- and Fourt...Med Sci Monit In Press; DOI: 10.12659/MSM.943468
08 Mar 2024 : Animal Research
Modification of Experimental Model of Necrotizing Enterocolitis (NEC) in Rat Pups by Single Exposure to Hyp...Med Sci Monit In Press; DOI: 10.12659/MSM.943443
18 Apr 2024 : Clinical Research
Comparative Analysis of Open and Closed Sphincterotomy for the Treatment of Chronic Anal Fissure: Safety an...Med Sci Monit In Press; DOI: 10.12659/MSM.944127
08 Mar 2024 : Laboratory Research
Evaluation of Retentive Strength of 50 Endodontically-Treated Single-Rooted Mandibular Second Premolars Res...Med Sci Monit In Press; DOI: 10.12659/MSM.944110
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952








