15 December 2020: Clinical Research
Measurement of Cyclin D2 () Gene Promoter Methylation in Plasma and Peripheral Blood Mononuclear Cells and Alpha-Fetoprotein Levels in Patients with Hepatitis B Virus-Associated Hepatocellular Carcinoma
Yu Qian1ABCDEF, Jing-Wen Wang1BC, Yu Fang1BCD, Xiao-Dong Yuan1B, Yu-Chen Fan12ADF, Shuai Gao12CDFG, Kai Wang12ADEG*DOI: 10.12659/MSM.927444
Med Sci Monit 2020; 26:e927444
Abstract
BACKGROUND: Alpha-fetoprotein (AFP) is widely used to screen for hepatocellular carcinoma (HCC). However, the use of this biomarker has been challenged due to its low sensitivity and high rate of false negatives. In this study, we evaluated the diagnostic capability of cyclin D2 (CCND2) promoter methylation in patients with HCC related to hepatitis B virus (HBV).
MATERIAL AND METHODS: Using methylation-specific PCR and quantitative real-time PCR, we measured methylation status and mRNA levels of CCND2 in plasma and peripheral blood mononuclear cells (PBMCs) from 275 subjects: 75 patients with chronic hepatitis B (CHB), 47 with liver cirrhosis (LC), 118 with HCC, and 35 healthy controls (HCs).
RESULTS: The methylation rate of the CCND2 promoter was significantly higher in HCC patients than in patients without HCC (P<0.001). Furthermore, advanced HCC (TNM III/IV) was associated with a significantly higher frequency of CCND2 methylation and lower CCND2 mRNA levels than early-stage disease (TNM I/II; P<0.05). Combined measurement of CCND2 methylation and AFP yielded significantly higher sensitivity and area under the curve (AUC) than AFP alone in distinguishing patients with HCC from subjects with LC and CHB (P<0.001).
CONCLUSIONS: CCND2 methylation may be useful for predicting HCC progression. In addition, combined measurement of CCND2 methylation and AFP could serve as a non-invasive diagnostic marker for patients with HBV-related HCC.
Keywords: Carcinoma, Hepatocellular, Cyclin D2, Diagnosis, DNA Methylation, Area Under Curve, Diagnosis, Differential, Disease Progression, Hepatitis B, Chronic, Leukocytes, Mononuclear, Liver Cirrhosis, Liver Neoplasms, Promoter Regions, Genetic, RNA, Messenger, Sensitivity and Specificity, alpha-Fetoproteins
Background
Hepatocellular carcinoma (HCC), which constitutes the majority of liver cancers, is the second most common cause of death from cancer around the world [1,2]. In China, HCC is mainly caused by chronic hepatitis B virus (HBV) infection [3]. Although a multitude of diagnostic and therapeutic modalities have been developed for HCC, the long-term prognosis remains poor, and 5 year survival rates are low [4]. A combination of imaging technologies such as ultrasonography (US) and determination of alpha-fetoprotein (AFP) levels is the most widely used method for detecting HCC. However, several studies have shown that low sensitivity and high rates of both false negatives and false positives limit the wide application of serum AFP levels as a marker [5,6]. Farinati et al. reported that only 54% of patients with HCC had abnormal serum AFP levels in a large multicentric survey [7]. AFP has also been questioned for its low specificity, because elevated AFP levels are also found in pregnant women and in patients with active hepatitis and embryonic carcinomas [8]. US-based detection is affected by several factors, including the professional expertise of operators, the physical status of the patient, the presence or absence of cirrhosis, and tumor size [9]. A previous study demonstrated that US alone has a sensitivity of 32% for the diagnosis of early-stage HCC [10]. In addition, US cannot be used to visualize adequately the liver in patients with a nodular liver and does not have the accuracy to distinguish HCC from other lesions [11,12]. Collectively, these parameters determine whether US can effectively detect HCC at the early stage. Potential alternative imaging modalities are unsuitable, as computed tomography (CT) scans and magnetic resonance imaging (MRI) cannot detect small HCC lesions [13]. Hence, effective and non-invasive biomarkers for diagnosis of HCC are urgently required.
DNA methylation plays crucial roles in the progression of several types of human cancer [14,15]. Moreover, changes in DNA methylation patterns are frequently observed in the early stages of disease; for example, Zhang et al. reported that altered DNA methylation could be detected 1–9 years before HCC itself [16]. Several genes have been implicated in the disease stage and clinical outcome of HCC, including
Cyclin D2 (CCND2) regulates cell-cycle progression [21] and inhibits cell growth. Accordingly, CCND2 levels are elevated in normal human cells under growth arrest. Aberrant expression of CCND2 affects cell-cycle progression, suggesting that CCND2 has an additional function that maintains the non-proliferative state [22–24]. This effect can be relieved by the inhibition of
Material and Methods
PATIENTS AND CONTROLS:
A total of 118 patients with HCC, 47 with liver cirrhosis (LC), 75 with chronic hepatitis B (CHB), and 35 healthy controls (HCs) were recruited in the Department of Hepatology, Qilu Hospital of Shandong University, from March 2018 to December 2019. All patients were HBsAg-positive. Figure 1 depicts the selection process. The Ethics Committee of Qilu Hospital of Shandong University approved the study protocol, and informed consent was obtained from all participants prior to the study.
DNA EXTRACTION FROM PLASMA AND PBMCS:
Citrate-anticoagulated peripheral blood (5 mL) was collected from all subjects. DNA was extracted from 400 μL plasma using the QIAamp DNA Blood Mini Kit (Qiagen, Mainz, Germany) and stored at −20°C until use. After centrifugation on a Ficoll-Paque Plus density gradient (GE Healthcare, Uppsala, Sweden), PBMCs were collected from the interface and washed 3 times with phosphate-buffered saline. Genomic DNA was extracted from PBMCs using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Extracted DNA was eluted in 200 μL sterile water and stored at −20°C until use.
SODIUM BISULFITE MODIFICATION AND MSP:
Extracted DNA was modified using the EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA, USA). Treatment with bisulfite converts unmethylated cytosine residues to uracil but does not affect methylated cytosine residues. Modified DNA was dissolved at a final volume of 20 μL and stored at −20°C until use. Bisulfite-modified DNA was amplified using methylated and unmethylated primers specific for the CCND2 promoter. The primer pairs used for MSP analysis of CCND2 were described previously [26].
Methylated sequence (276 bp PCR product):
Unmethylated sequence (222 bp PCR product):
The MSP reaction mixture had a volume of 25 μL as follows: 10.5 μL nuclease-free water, 12.5 μL PreMix Taq (Zymo Research, Irvine, CA, USA), 0.5 μL of each primer (10 μmol/L), and 1 μL bisulfite-treated DNA. Touchdown PCR conditions were as follows: 95°C for 5 min; ten cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 45 s; 30 cycles of 95°C for 30 s, 45°C for 30 s, and 72°C for 45 s; and a final extension step at 72°C for 5 min. Water without DNA was used as a negative control. PCR products (7 μL) were electrophoresed on 2% agarose gels and visualized under UV illumination after staining with Gel Red (Biotium, Fremont, CA, USA).
RNA EXTRACTION FROM PBMCS AND QUANTITATIVE REAL-TIME PCR (QRT-PCR):
RNA was extracted from PBMCs by the phenol–chloroform–isopropanol method. Total RNA was resuspended in 20 μL RNase-free water. Subsequently, RNA was converted into cDNA using the PrimeScript RT Reagent Kit (Takara, Shiga, Japan).
Expression of CCND2 mRNA was detected by qRT-PCR on an Agilent Technologies Stratagene Mx3005P (Stratagene, La Jolla, CA, USA) using SYBR Green PCR Mix (Takara, Shiga, Japan). ACTB (encoding β-actin) was used as an endogenous control. The primer pairs used for qRT-PCR analysis of CCND2 [25] and ACTB [19] were described previously. qRT-PCR was performed as follows: initial denaturation at 95°C for 30 s, followed by 40 cycles of 95°C for 5 s, 60°C for 30 s, 72°C for 30 s, 65°C for 15 s, and 40°C for 30 s. mRNA levels were calculated using the comparative (2−ΔΔCt) method.
STATISTICAL ANALYSIS:
All data were analyzed using SPSS 17.0 software (SPSS Inc., Chicago, IL, USA). The Mann-Whitney U-test was used to compare
Results
GENERAL CHARACTERISTICS:
Clinicopathological characteristics of the participants are provided in Table 1.
CCND2 MRNA LEVELS IN PBMCS OF HCC:
CCND2 mRNA levels were significantly lower in HCC patients than in LC patients (P<0.001), CHB patients (P<0.001), and HCs (P<0.001; Figure 2A). However, there were no significant differences in CCND2 mRNA levels between LC patients, CHB patients, and HCs (P>0.05, respectively).
Moreover, HCC patients with advanced disease (TNM stage III/IV) had significantly lower CCND2 mRNA levels than patients with early-stage disease (TNM I/II; P=0.030; Figure 2B). CCND2 mRNA levels in the HCC group exhibited no significant relationship with age, HBeAg, gender, smoking status, alcohol use, AFP, tumor number, tumor size, vascular invasion, or CTP staging (Figure 2C–2L).
METHYLATION FREQUENCY OF CCND2 AND ITS CORRELATION WITH GENE TRANSCRIPTION:
The frequency of CCND2 promoter methylation was significantly higher in HCC patients (55/118, 46.61% in PBMCs; 52/118, 44.07% in plasma) than in LC patients (8/47, 17.02% in PBMCs; 5/47, 10.64% in plasma; P<0.001), CHB patients (8/75, 10.67% in PBMCs; 7/75, 9.33% in plasma; P<0.001), and HCs (4/35, 11.43% in PBMCs; 2/35, 5.71% in plasma; P<0.001; Figure 3A, 3B). However, CCND2 methylation frequencies did not differ significantly between LC patients, CHB patients, and HCs (P>0.05, respectively).
To determine whether altered promoter methylation could affect CCND2 transcription, we compared CCND2 mRNA levels in subjects with and without promoter methylation. In the HCC group, the level of CCND2 mRNA was significantly lower in methylated subjects than in unmethylated subjects (P=0.041; Figure 3C). These data support our hypothesis. Figure 3D shows a representative result of agarose gel electrophoresis.
ASSOCIATION BETWEEN CCND2 PROMOTER METHYLATION AND HCC PROGRESSION:
Table 2 shows that the methylation frequency of the CCND2 promoter in HCC patients was significantly higher in HCC patients with vascular invasion than in those without vascular invasion (P=0.012 in PBMCs; P=0.008 in plasma). In addition, CCND2 promoter hypermethylation was more common in HCC patients with advanced disease (TNM III/IV) than in those with early-stage disease (TNM I/II; P=0.004 in PBMCs; P=0.001 in plasma). The CCND2 methylation rate increased gradually with TNM stage (Table 3). Together, these results reveal that CCND2 methylation is more frequent in advanced-stage HCC patients. We found no significant correlations between CCND2 methylation status and other parameters.
DIAGNOSTIC UTILITY OF CCND2 PROMOTER METHYLATION AND AFP LEVEL:
For discrimination of HCC from LC, CCND2 promoter methylation had a sensitivity of 46.61% in PBMCs and 44.07% in plasma, and a specificity of 82.98% in PBMCs and 89.36% in plasma. For discrimination of HCC from CHB, CCND2 promoter methylation had a sensitivity of 46.61% in PBMCs and 44.07% in plasma, and a specificity of 89.33% in PBMCs and 90.67% in plasma (Table 4). Figure 4A and 4B show that the AUC of combined measurement of CCND2 promoter methylation and AFP level was significantly higher than that of AFP alone (0.698 vs. 0.540 in PBMCs, P<0.001; 0.694 vs. 0.540 in plasma, P<0.001) in discriminating HCC from LC. The AUC of combined measurement was also significantly higher than that of AFP alone (0.724 vs. 0.571 in PBMCs, P<0.001; 0.720 vs. 0.571 in plasma, P<0.001) in discriminating HCC from CHB (Figure 4C, 4D).
Next, we compared the diagnostic value of combined measurement of CCND2 methylation and AFP with that of AFP alone for discriminating HCC from LC. As shown in Figure 5, the HCC detection rate in the CCND2-methylated group was significantly higher than that in the CCND2-unmethylated group, regardless of whether the AFP level was ≤20 ng/mL or >20 ng/mL (χ2=14.246, P<0.001, AFP ≤20 ng/mL; χ2=5.499, P=0.018, AFP >20 ng/mL). We defined AFP >20 ng/mL or the presence of CCND2 methylation as positive. Table 4 revealed that for discrimination of HCC from LC, measurement of CCND2 methylation plus AFP had a sensitivity of 82.20% (97/118) in PBMCs and 81.36% (96/118) in plasma; a specificity of 57.45% (27/47) in both PBMCs and plasma; a PPV of 82.91% (97/117) in PBMCs and 82.76% (96/116) in plasma; and a negative predictive value (NPV) of 56.25% (27/48) in PBMCs and 55.10% (27/49) in plasma. Notably, the sensitivity of combined measurement was significantly higher than that of AFP alone (P<0.001). Similarly, for discriminating HCC from CHB, combined measurement had higher sensitivity (82.20% vs. 57.63% in PBMCs, P<0.001; 81.36% vs. 57.63% in plasma, P<0.001) and NPV (69.12% vs. 48.98% in PBMCs, P=0.010; 68.12% vs. 48.98% in plasma, P=0.014) than AFP alone (Table 4).
Discussion
CCND2 is involved in cell-cycle regulation, in which its critical function involves the formation of a complex with subunits of CDK6 and CDK4, resulting in phosphorylation of retinoblastoma protein (RB) [21,26]. Overexpression of CCND2 is correlated with progression and poor prognosis of several cancers, indicating that CCND2 should be considered as a proto-oncogene [29,30]. However, silencing of CCND2 expression by promoter methylation is associated with cancer progression, indicating that
More importantly,
Currently, AFP is the biomarker used most widely for HCC screening and diagnosis. However, the sensitivity of serum AFP is only 22–60%, depending on the cut-off point [7,37,38]; consequently, its clinical value is limited. As shown in Figure 5, the rate of HCC detection was significantly higher in the
Several studies showed that PBMCs and plasma cell-free DNA are useful for identifying gene mutation and abnormal DNA methylation, and can therefore be used for diagnosis of human cancers [39–41]. In this study, we measured
One weakness of this study is that the MSP method is a qualitative approach for detecting gene methylation. Other methods, such as direct sequencing, may provide more accurate information about methylation. However, MSP can be performed rapidly and easily, enabling most clinical laboratories to carry it out. Although the results of MSP do not provide complete information, the method can still be used to select methylated subjects that may require further examination by sequencing. PIVKA-II, a prothrombin induced by vitamin K deficiency, is increased in malignant hepatocytes [43], suggesting that it may have potential as a biomarker for HCC. A number of studies have revealed that combined determination of PIVKA-II and AFP can improve the diagnostic accuracy of HCC detection compared with either of these biomarkers alone [44,45]. In the present study, we did not assess the diagnostic value of PIVKA-II alone or in combination with
Conclusions
Methylation of the
Figures
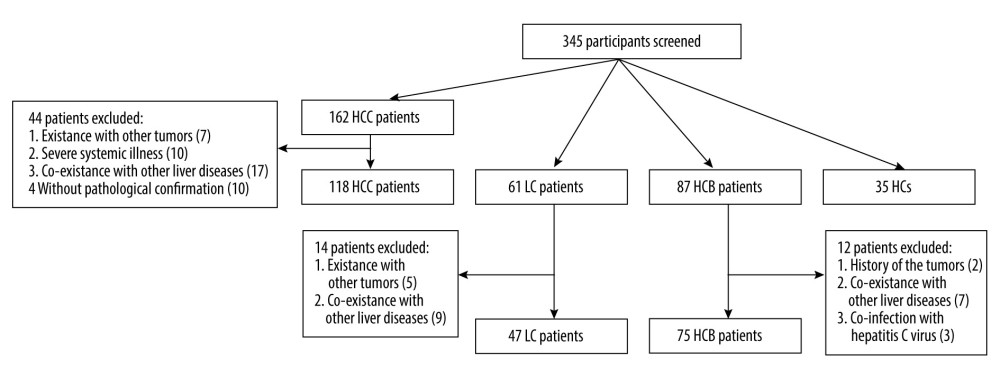 Figure 1. Flowchart describing the participant selection process. HCC – hepatocellular carcinoma; LC – liver cirrhosis; CHB – chronic hepatitis B; HCs – healthy controls.
Figure 1. Flowchart describing the participant selection process. HCC – hepatocellular carcinoma; LC – liver cirrhosis; CHB – chronic hepatitis B; HCs – healthy controls. 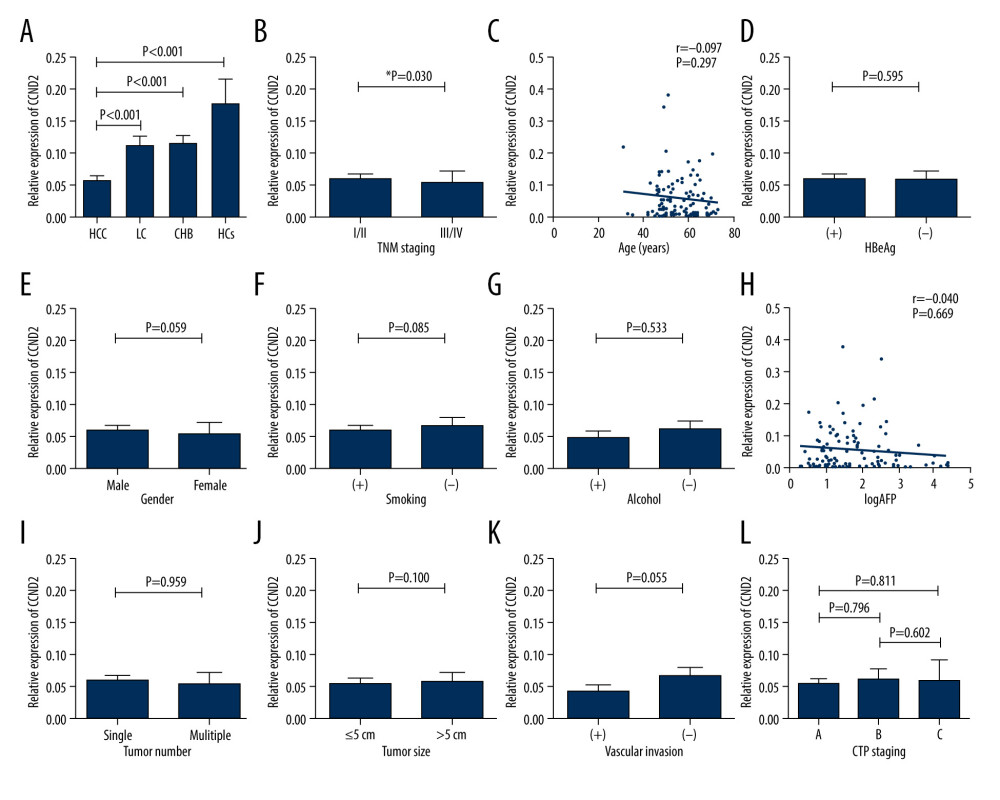 Figure 2. CCND2 mRNA level in the indicated groups and its correlations with clinical characteristics of HCC. (A) CCND2 mRNA levels differed significantly among the 4 groups (HCC vs. LC, P<0.001; HCC vs. CHB, P<0.001; HCC vs. HCs, P<0.001). (B) mRNA levels in HCC patients differed significantly between advanced tumor node metastasis (TNM) stage (III/IV) and early TNM stage (I/II; P=0.030). (C) mRNA level was not correlated with age in 118 HCC patients (P=0.297). (D–G) mRNA levels in HCC patients did not significantly differ between HBeAg-positive and HBeAg-negative patients (P=0.595), male and female patients (P=0.059), smokers and non-smokers (P=0.085), or drinkers and non-drinkers (P=0.533). (H) mRNA levels and serum AFP levels were not significantly correlated in HCC patients (P=0.669). (I–K) mRNA levels did not significantly differ between HCC patients with a single tumor and those with multiple tumors (P=0.959), those with tumors ≤5 cm and those with tumors >5 cm (P=0.100), or HCC patients with and without vascular invasion (P=0.055). (L) mRNA levels did not differ among CTP stages within the HCC group (A vs. B, P=0.796; A vs. C, P=0.811; B vs. C, P=0.602).
Figure 2. CCND2 mRNA level in the indicated groups and its correlations with clinical characteristics of HCC. (A) CCND2 mRNA levels differed significantly among the 4 groups (HCC vs. LC, P<0.001; HCC vs. CHB, P<0.001; HCC vs. HCs, P<0.001). (B) mRNA levels in HCC patients differed significantly between advanced tumor node metastasis (TNM) stage (III/IV) and early TNM stage (I/II; P=0.030). (C) mRNA level was not correlated with age in 118 HCC patients (P=0.297). (D–G) mRNA levels in HCC patients did not significantly differ between HBeAg-positive and HBeAg-negative patients (P=0.595), male and female patients (P=0.059), smokers and non-smokers (P=0.085), or drinkers and non-drinkers (P=0.533). (H) mRNA levels and serum AFP levels were not significantly correlated in HCC patients (P=0.669). (I–K) mRNA levels did not significantly differ between HCC patients with a single tumor and those with multiple tumors (P=0.959), those with tumors ≤5 cm and those with tumors >5 cm (P=0.100), or HCC patients with and without vascular invasion (P=0.055). (L) mRNA levels did not differ among CTP stages within the HCC group (A vs. B, P=0.796; A vs. C, P=0.811; B vs. C, P=0.602). 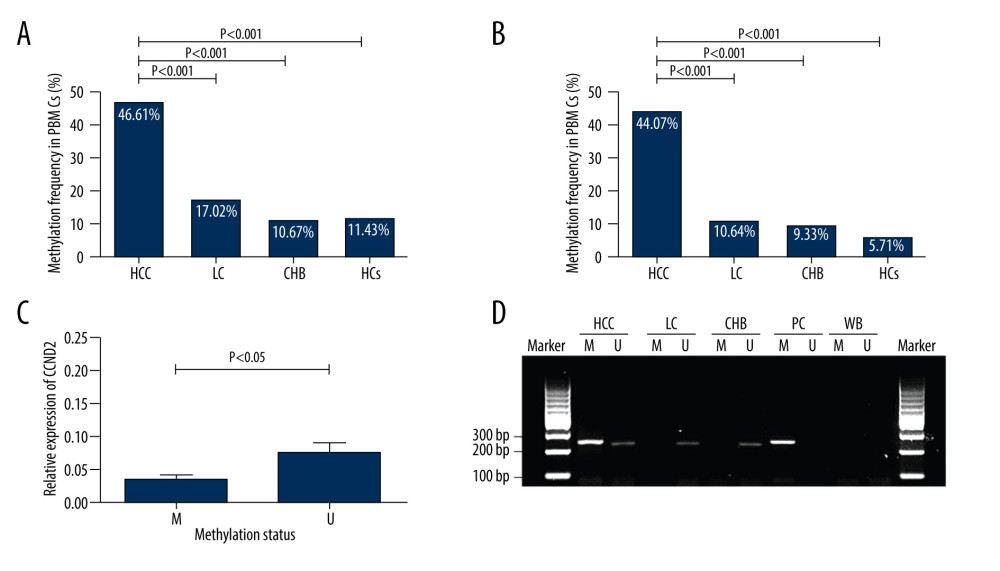 Figure 3. Methylation status of CCND2 promoter and correlation with expression of CCND2 mRNA. (A) In total, 55/118 (46.61%) hepatocellular carcinoma (HCC) patients, 8/47 (17.02%) liver cirrhosis (LC) patients, 8/75 (10.67%) chronic hepatitis B (CHB) patients, and 4/35 (11.43%) healthy controls (HCs) exhibited aberrant CCND2 promoter methylation in PBMCs. (B) 52/118 (44.07%) HCC patients, 5/47 (10.64%) LC patients, 7/75 (9.33%) CHB patients, and 2/35 (5.71%) HCs exhibited aberrant CCND2 promoter methylation in plasma. (C) CCND2 mRNA levels differed significantly between the methylation group and non-methylation groups (P<0.05). (D) Representative measurements of CCND2 promoter by methylation-specific polymerase chain reaction (MSP). PC – positive control; WB – water blank; M – methylated sequence; U – unmethylated sequence.
Figure 3. Methylation status of CCND2 promoter and correlation with expression of CCND2 mRNA. (A) In total, 55/118 (46.61%) hepatocellular carcinoma (HCC) patients, 8/47 (17.02%) liver cirrhosis (LC) patients, 8/75 (10.67%) chronic hepatitis B (CHB) patients, and 4/35 (11.43%) healthy controls (HCs) exhibited aberrant CCND2 promoter methylation in PBMCs. (B) 52/118 (44.07%) HCC patients, 5/47 (10.64%) LC patients, 7/75 (9.33%) CHB patients, and 2/35 (5.71%) HCs exhibited aberrant CCND2 promoter methylation in plasma. (C) CCND2 mRNA levels differed significantly between the methylation group and non-methylation groups (P<0.05). (D) Representative measurements of CCND2 promoter by methylation-specific polymerase chain reaction (MSP). PC – positive control; WB – water blank; M – methylated sequence; U – unmethylated sequence. 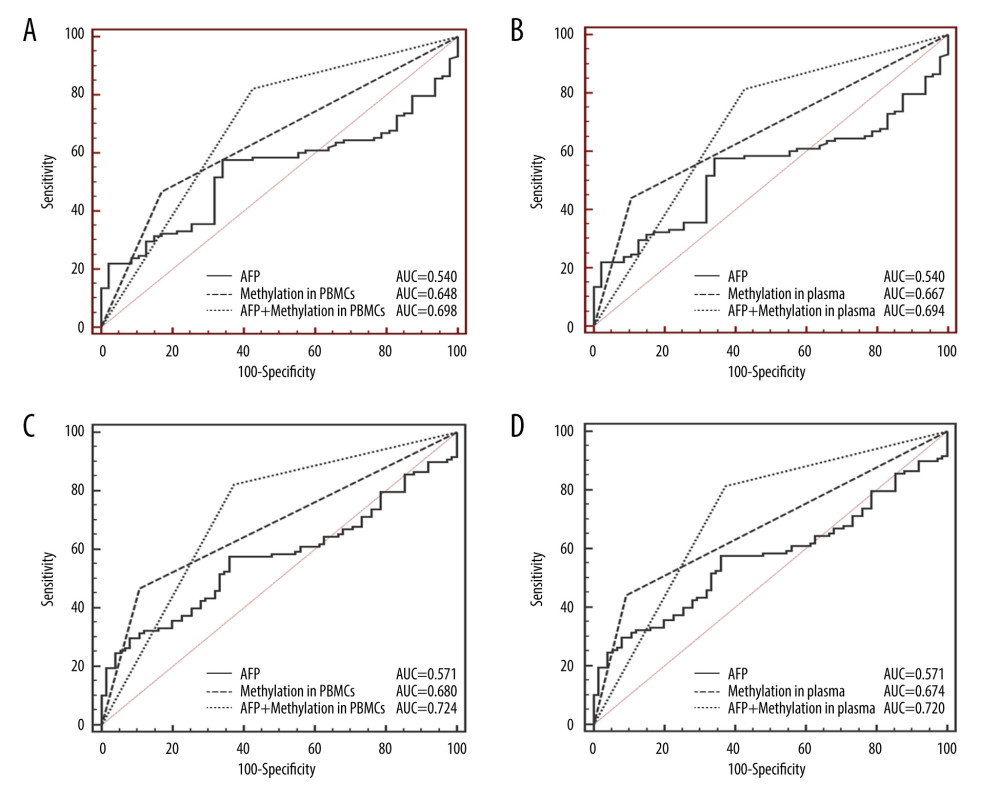 Figure 4. Receiver operating characteristic curves (ROC) of AFP, CCND2 promoter methylation, and combined measurement for distinguishing HCC from LC and CHB in both PBMCs and plasma. (A) The area under the ROC curves (AUC) of combined measurement in PBMCs was significantly higher than that of AFP levels for discriminating HCC from LC patients (AUC 0.698 vs. 0.540, P<0.001). (B) The AUC of combined measurement in plasma was significantly higher than that of AFP levels for discriminating HCC from LC patients (AUC 0.694 vs. 0.540, P<0.001). (C) The AUC of combined measurement in PBMCs was significantly higher than that of AFP levels for discriminating HCC from CHB patients (AUC 0.724 vs. 0.571, P<0.001). (D) The AUC of combined measurement in plasma was significantly higher than that of AFP levels for discriminating HCC from CHB patients (AUC 0.720 vs. 0.571, P<0.001).
Figure 4. Receiver operating characteristic curves (ROC) of AFP, CCND2 promoter methylation, and combined measurement for distinguishing HCC from LC and CHB in both PBMCs and plasma. (A) The area under the ROC curves (AUC) of combined measurement in PBMCs was significantly higher than that of AFP levels for discriminating HCC from LC patients (AUC 0.698 vs. 0.540, P<0.001). (B) The AUC of combined measurement in plasma was significantly higher than that of AFP levels for discriminating HCC from LC patients (AUC 0.694 vs. 0.540, P<0.001). (C) The AUC of combined measurement in PBMCs was significantly higher than that of AFP levels for discriminating HCC from CHB patients (AUC 0.724 vs. 0.571, P<0.001). (D) The AUC of combined measurement in plasma was significantly higher than that of AFP levels for discriminating HCC from CHB patients (AUC 0.720 vs. 0.571, P<0.001). 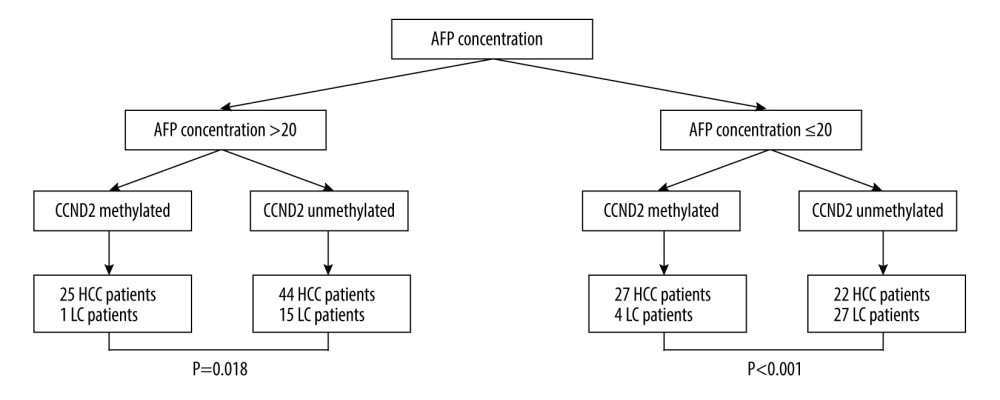 Figure 5. Classification of 118 HCC and 47 LC patients according to AFP concentrations and CCND2 methylation status in plasma. AFP – alpha-fetoprotein; HCC – hepatocellular carcinoma; LC – liver cirrhosis.
Figure 5. Classification of 118 HCC and 47 LC patients according to AFP concentrations and CCND2 methylation status in plasma. AFP – alpha-fetoprotein; HCC – hepatocellular carcinoma; LC – liver cirrhosis. Tables
Table 1. Characterization of the study participants.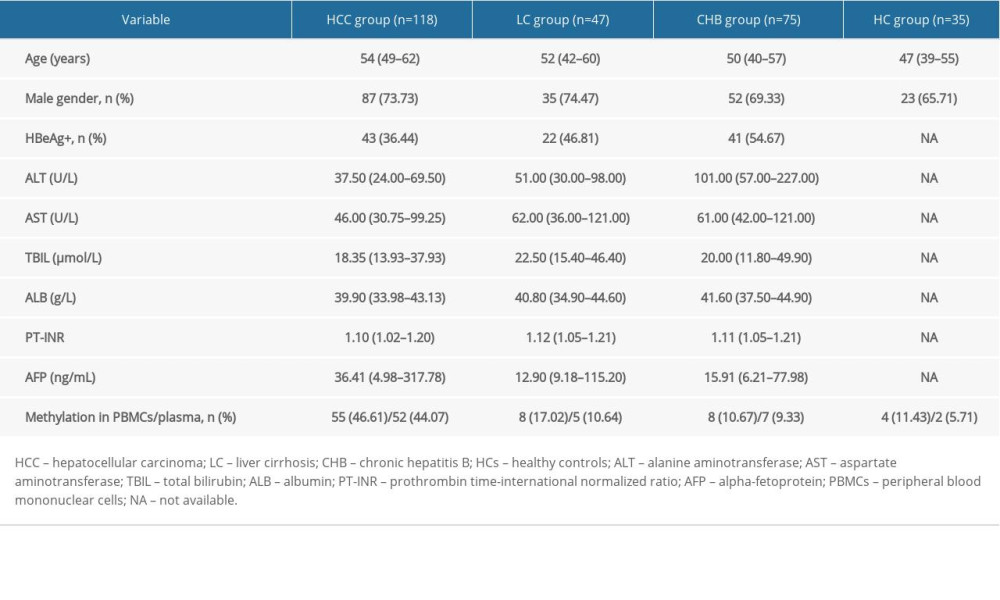 Table 2. Association between CCND2 methylation status and clinical characteristics of HCC patients.
Table 2. Association between CCND2 methylation status and clinical characteristics of HCC patients.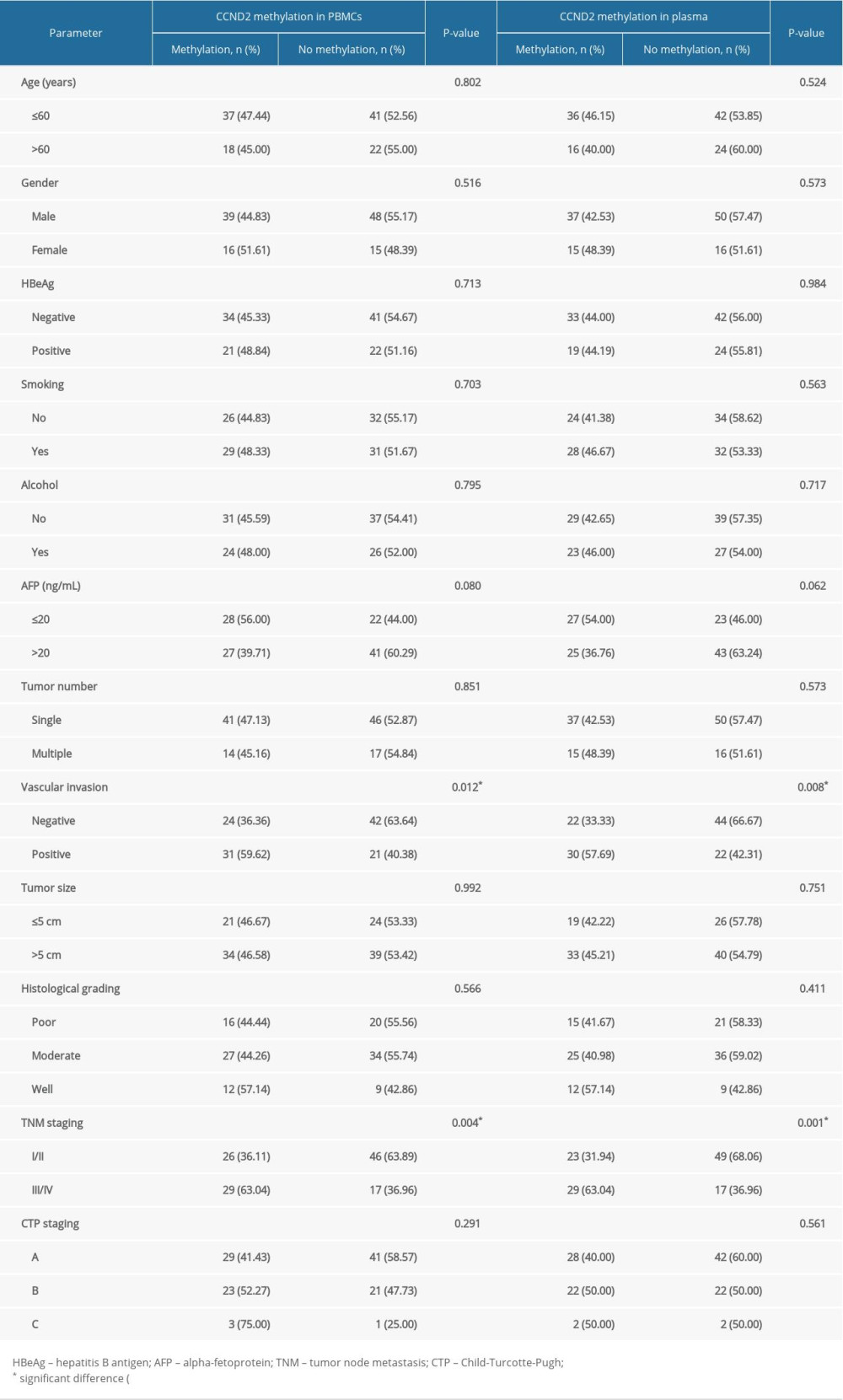 Table 3. CCND2 methylation levels in PBMCs and TNM stage from HCC patients.
Table 3. CCND2 methylation levels in PBMCs and TNM stage from HCC patients.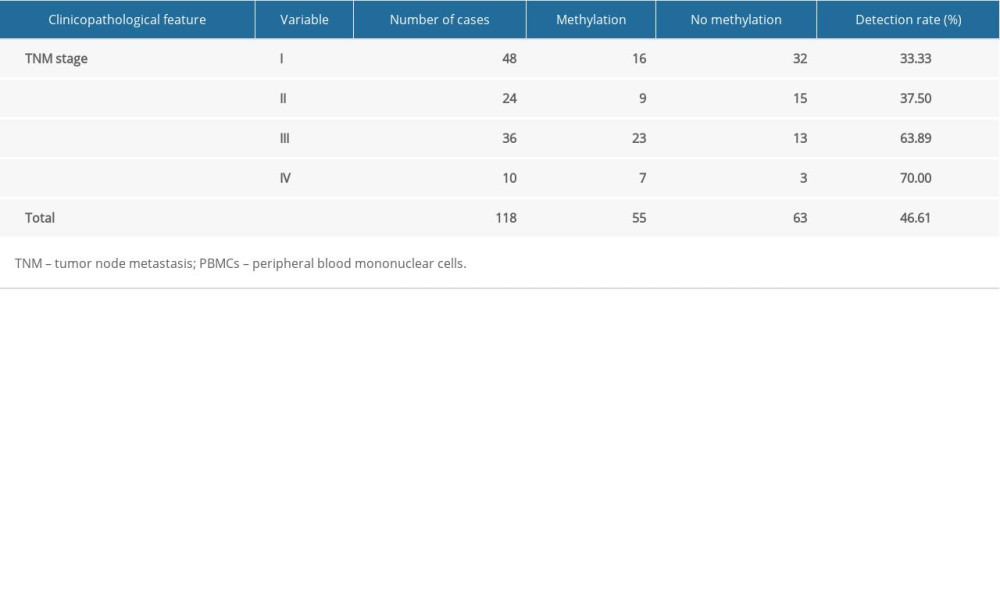 Table 4. Diagnostic utility of AFP, CCND2 promoter methylation, and combined measurement for discrimination of HCC from LC and CHB.
Table 4. Diagnostic utility of AFP, CCND2 promoter methylation, and combined measurement for discrimination of HCC from LC and CHB.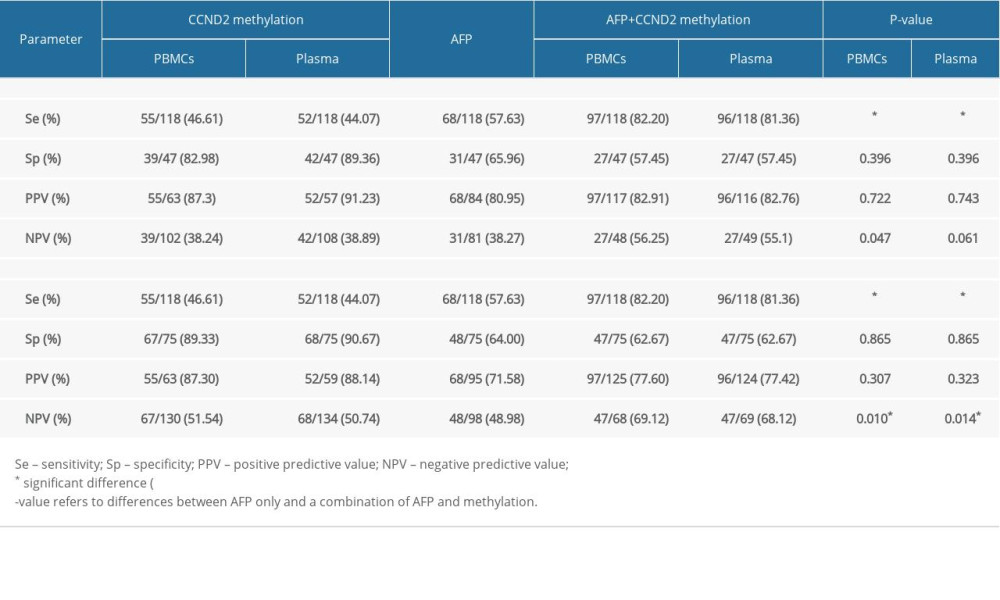
References
1. Torre LA, Bray F, Siegel RL, Global cancer statistics, 2012: Cancer J Clin, 2015; 65(2); 87-108
2. Zhu RX, Seto WK, Lai CL, Yuen MF, Epidemiology of hepatocellular carcinoma in the Asia-Pacific Region: Gut Liver, 2016; 10(3); 332-39
3. Chen W, Zheng R, Baade PD, Cancer statistics in China, 2015: Cancer J Clin, 2016; 66(2); 115-32
4. Fong ZV, Tanabe KK, The clinical management of hepatocellular carcinoma in the United States, Europe, and Asia: A comprehensive and evidence-based comparison and review: Cancer, 2014; 120(18); 2824-38
5. Villanueva A, Minguez B, Forner A, Hepatocellular carcinoma: Novel molecular approaches for diagnosis, prognosis, and therapy: Ann Rev Med, 2010; 61; 317-28
6. Huang TS, Shyu YC, Turner R, Diagnostic performance of alpha-fetoprotein, lens culinaris agglutinin-reactive alpha-fetoprotein, des-gamma carboxyprothrombin, and glypican-3 for the detection of hepatocellular carcinoma: A systematic review and meta-analysis protocol: Syst Rev, 2013; 2; 37
7. Farinati F, Marino D, De Giorgio M, Diagnostic and prognostic role of alpha-fetoprotein in hepatocellular carcinoma: both or neither?: Am J Gastroenterol, 2006; 101(3); 524-32
8. Forner A, Reig M, Bruix J, Hepatocellular carcinoma: Lancet (London, England), 2018; 391(10127); 1301-14
9. Singal AG, Conjeevaram HS, Volk ML, Effectiveness of hepatocellular carcinoma surveillance in patients with cirrhosis: Cancer Epidemiol Biomarker Prev, 2012; 21(5); 793-99
10. Singal A, Volk ML, Waljee A, Meta-analysis: Surveillance with ultrasound for early-stage hepatocellular carcinoma in patients with cirrhosis: Aliment Pharmacol Ther, 2009; 30(1); 37-47
11. Simmons O, Fetzer DT, Yokoo T, Predictors of adequate ultrasound quality for hepatocellular carcinoma surveillance in patients with cirrhosis: Aliment Pharmacol Ther, 2017; 45(1); 169-77
12. Chon YE, Jung KS, Kim MJ, Predictors of failure to detect early hepatocellular carcinoma in patients with chronic hepatitis B who received regular surveillance: Aliment Pharmacol Ther, 2018; 47(8); 1201-12
13. Omata M, Lesmana LA, Tateishi R, Asian Pacific Association for the Study of the Liver consensus recommendations on hepatocellular carcinoma: Hepatol Int, 2010; 4(2); 439-74
14. Sundar IK, Yin Q, Baier BS, DNA methylation profiling in peripheral lung tissues of smokers and patients with COPD: Clin Epigenetics, 2017; 9; 38
15. Wu HC, Southey MC, Hibshoosh H, DNA methylation in breast tumor from high-risk women in the breast cancer family registry: Anticancer Res, 2017; 37(2); 659-64
16. Zhang YJ, Wu HC, Shen J, Predicting hepatocellular carcinoma by detection of aberrant promoter methylation in serum DNA: Clin Cancer Res, 2007; 13(8); 2378-84
17. Feng Q, Stern JE, Hawes SE, DNA methylation changes in normal liver tissues and hepatocellular carcinoma with different viral infection: Exp Mol Pathol, 2010; 88(2); 287-92
18. Nishida N, Arizumi T, Takita M, Quantification of tumor DNA in serum and vascular invasion of human hepatocellular carcinoma: Oncology, 2013; 84(Suppl 1); 82-87
19. Li F, Fan YC, Gao S, Methylation of serum insulin-like growth factor-binding protein 7 promoter in hepatitis B virus-associated hepatocellular carcinoma: Genes Chromosomes Cancer, 2014; 53(1); 90-97
20. Wang J, Qin Y, Li B, Detection of aberrant promoter methylation of GSTP1 in the tumor and serum of Chinese human primary hepatocellular carcinoma patients: Clin Biochem, 2006; 39(4); 344-48
21. Sherr CJ, D-type cyclins: Trends Biochem Sci, 1995; 20(5); 187-90
22. Tam SW, Theodoras AM, Shay JW, Differential expression and regulation of Cyclin D1 protein in normal and tumor human cells: Association with Cdk4 is required for Cyclin D1 function in G1 progression: Oncogene, 1994; 9(9); 2663-74
23. Meyyappan M, Wong H, Hull C, Riabowol KT, Increased expression of cyclin D2 during multiple states of growth arrest in primary and established cells: Mol Cell Biol, 1998; 18(6); 3163-72
24. Meyyappan M, Atadja PW, Riabowol KT, Regulation of gene expression and transcription factor binding activity during cellular aging: Biol Signals, 1996; 5(3); 130-38
25. Tsutsui M, Iizuka N, Moribe T, Methylated cyclin D2 gene circulating in the blood as a prognosis predictor of hepatocellular carcinoma: Clin Chim Acta, 2010; 411(7–8); 516-20
26. Evron E, Umbricht CB, Korz D, Loss of cyclin D2 expression in the majority of breast cancers is associated with promoter hypermethylation: Cancer Res, 2001; 61(6); 2782-87
27. Yu J, Leung WK, Ebert MP, Absence of cyclin D2 expression is associated with promoter hypermethylation in gastric cancer: Br J Cancer, 2003; 88(10); 1560-65
28. Hsu A, Wong CP, Yu Z, Promoter de-methylation of cyclin D2 by sulforaphane in prostate cancer cells: Clin Epigenetics, 2011; 3(1); 3
29. Mermelshtein A, Gerson A, Walfisch S, Expression of D-type cyclins in colon cancer and in cell lines from colon carcinomas: Br J Ccancer, 2005; 93(3); 338-45
30. Dhillon VS, Shahid M, Husain SA, CpG methylation of the FHIT, FANCF, cyclin-D2, BRCA2 and RUNX3 genes in Granulosa cell tumors (GCTs) of ovarian origin: Mol Cancer, 2004; 3; 33
31. Matsubayashi H, Sato N, Fukushima N, Methylation of cyclin D2 is observed frequently in pancreatic cancer but is also an age-related phenomenon in gastrointestinal tissues: Clin Cancer Res, 2003; 9(4); 1446-52
32. Tahara T, Arisawa T, DNA methylation as a molecular biomarker in gastric cancer: Epigenomics, 2015; 7(3); 475-86
33. Henrique R, Costa VL, Cerveira N, Hypermethylation of Cyclin D2 is associated with loss of mRNA expression and tumor development in prostate cancer: J Mol Med (Berl), 2006; 84(11); 911-18
34. Callahan CL, Wang Y, Marian C, DNA methylation and breast tumor clinicopathological features: The Western New York Exposures and Breast Cancer (WEB) study: Epigenetics, 2016; 11(9); 643-52
35. Lehmann U, Langer F, Feist H, Quantitative assessment of promoter hypermethylation during breast cancer development: Am J Pathol, 2002; 160(2); 605-12
36. Padar A, Sathyanarayana UG, Suzuki M, Inactivation of cyclin D2 gene in prostate cancers by aberrant promoter methylation: Clin Cancer Res, 2003; 9(13); 4730-34
37. Lok AS, Sterling RK, Everhart JE, Des-gamma-carboxy prothrombin and alpha-fetoprotein as biomarkers for the early detection of hepatocellular carcinoma: Gastroenterology, 2010; 138(2); 493-502
38. Marrero JA, Feng Z, Wang Y, Alpha-fetoprotein, des-gamma carboxyprothrombin, and lectin-bound alpha-fetoprotein in early hepatocellular carcinoma: Gastroenterology, 2009; 137(1); 110-18
39. Terry MB, Delgado-Cruzata L, Vin-Raviv N, DNA methylation in white blood cells: Association with risk factors in epidemiologic studies: Epigenetics, 2011; 6(7); 828-37
40. Friso S, Udali S, Guarini P, Global DNA hypomethylation in peripheral blood mononuclear cells as a biomarker of cancer risk: Cancer Epidemiol Biomarkers Prev, 2013; 22(3); 348-55
41. Pezzuto F, Buonaguro L, Buonaguro FM, Tornesello ML, The role of circulating free DNA and microRNA in non-invasive diagnosis of HBV- and HCV-related hepatocellular carcinoma: Int J Mol Sci, 2018; 19(4); 1007
42. Iizuka N, Oka M, Yamada-Okabe H, Comparison of gene expression profiles between hepatitis B virus- and hepatitis C virus-infected hepatocellular carcinoma by oligonucleotide microarray data on the basis of a supervised learning method: Cancer Res, 2002; 62(14); 3939-44
43. Xing H, Yan C, Cheng L, Clinical application of protein induced by vitamin K antagonist-II as a biomarker in hepatocellular carcinoma: Tumour Biol, 2016 [Online ahead of print]
44. Wang Q, Chen Q, Zhang X, Diagnostic value of gamma-glutamyltransferase/aspartate aminotransferase ratio, protein induced by vitamin K absence or antagonist II, and alpha-fetoprotein in hepatitis B virus-related hepatocellular carcinoma: World J Gastroenterol, 2019; 25(36); 5515-29
45. Loglio A, Iavarone M, Facchetti F, The combination of PIVKA-II and AFP improves the detection accuracy for HCC in HBV caucasian cirrhotics on long-term oral therapy: Liver Int, 2020 [Online ahead of print]
Figures
 Figure 1. Flowchart describing the participant selection process. HCC – hepatocellular carcinoma; LC – liver cirrhosis; CHB – chronic hepatitis B; HCs – healthy controls.
Figure 1. Flowchart describing the participant selection process. HCC – hepatocellular carcinoma; LC – liver cirrhosis; CHB – chronic hepatitis B; HCs – healthy controls. Figure 2. CCND2 mRNA level in the indicated groups and its correlations with clinical characteristics of HCC. (A) CCND2 mRNA levels differed significantly among the 4 groups (HCC vs. LC, P<0.001; HCC vs. CHB, P<0.001; HCC vs. HCs, P<0.001). (B) mRNA levels in HCC patients differed significantly between advanced tumor node metastasis (TNM) stage (III/IV) and early TNM stage (I/II; P=0.030). (C) mRNA level was not correlated with age in 118 HCC patients (P=0.297). (D–G) mRNA levels in HCC patients did not significantly differ between HBeAg-positive and HBeAg-negative patients (P=0.595), male and female patients (P=0.059), smokers and non-smokers (P=0.085), or drinkers and non-drinkers (P=0.533). (H) mRNA levels and serum AFP levels were not significantly correlated in HCC patients (P=0.669). (I–K) mRNA levels did not significantly differ between HCC patients with a single tumor and those with multiple tumors (P=0.959), those with tumors ≤5 cm and those with tumors >5 cm (P=0.100), or HCC patients with and without vascular invasion (P=0.055). (L) mRNA levels did not differ among CTP stages within the HCC group (A vs. B, P=0.796; A vs. C, P=0.811; B vs. C, P=0.602).
Figure 2. CCND2 mRNA level in the indicated groups and its correlations with clinical characteristics of HCC. (A) CCND2 mRNA levels differed significantly among the 4 groups (HCC vs. LC, P<0.001; HCC vs. CHB, P<0.001; HCC vs. HCs, P<0.001). (B) mRNA levels in HCC patients differed significantly between advanced tumor node metastasis (TNM) stage (III/IV) and early TNM stage (I/II; P=0.030). (C) mRNA level was not correlated with age in 118 HCC patients (P=0.297). (D–G) mRNA levels in HCC patients did not significantly differ between HBeAg-positive and HBeAg-negative patients (P=0.595), male and female patients (P=0.059), smokers and non-smokers (P=0.085), or drinkers and non-drinkers (P=0.533). (H) mRNA levels and serum AFP levels were not significantly correlated in HCC patients (P=0.669). (I–K) mRNA levels did not significantly differ between HCC patients with a single tumor and those with multiple tumors (P=0.959), those with tumors ≤5 cm and those with tumors >5 cm (P=0.100), or HCC patients with and without vascular invasion (P=0.055). (L) mRNA levels did not differ among CTP stages within the HCC group (A vs. B, P=0.796; A vs. C, P=0.811; B vs. C, P=0.602). Figure 3. Methylation status of CCND2 promoter and correlation with expression of CCND2 mRNA. (A) In total, 55/118 (46.61%) hepatocellular carcinoma (HCC) patients, 8/47 (17.02%) liver cirrhosis (LC) patients, 8/75 (10.67%) chronic hepatitis B (CHB) patients, and 4/35 (11.43%) healthy controls (HCs) exhibited aberrant CCND2 promoter methylation in PBMCs. (B) 52/118 (44.07%) HCC patients, 5/47 (10.64%) LC patients, 7/75 (9.33%) CHB patients, and 2/35 (5.71%) HCs exhibited aberrant CCND2 promoter methylation in plasma. (C) CCND2 mRNA levels differed significantly between the methylation group and non-methylation groups (P<0.05). (D) Representative measurements of CCND2 promoter by methylation-specific polymerase chain reaction (MSP). PC – positive control; WB – water blank; M – methylated sequence; U – unmethylated sequence.
Figure 3. Methylation status of CCND2 promoter and correlation with expression of CCND2 mRNA. (A) In total, 55/118 (46.61%) hepatocellular carcinoma (HCC) patients, 8/47 (17.02%) liver cirrhosis (LC) patients, 8/75 (10.67%) chronic hepatitis B (CHB) patients, and 4/35 (11.43%) healthy controls (HCs) exhibited aberrant CCND2 promoter methylation in PBMCs. (B) 52/118 (44.07%) HCC patients, 5/47 (10.64%) LC patients, 7/75 (9.33%) CHB patients, and 2/35 (5.71%) HCs exhibited aberrant CCND2 promoter methylation in plasma. (C) CCND2 mRNA levels differed significantly between the methylation group and non-methylation groups (P<0.05). (D) Representative measurements of CCND2 promoter by methylation-specific polymerase chain reaction (MSP). PC – positive control; WB – water blank; M – methylated sequence; U – unmethylated sequence. Figure 4. Receiver operating characteristic curves (ROC) of AFP, CCND2 promoter methylation, and combined measurement for distinguishing HCC from LC and CHB in both PBMCs and plasma. (A) The area under the ROC curves (AUC) of combined measurement in PBMCs was significantly higher than that of AFP levels for discriminating HCC from LC patients (AUC 0.698 vs. 0.540, P<0.001). (B) The AUC of combined measurement in plasma was significantly higher than that of AFP levels for discriminating HCC from LC patients (AUC 0.694 vs. 0.540, P<0.001). (C) The AUC of combined measurement in PBMCs was significantly higher than that of AFP levels for discriminating HCC from CHB patients (AUC 0.724 vs. 0.571, P<0.001). (D) The AUC of combined measurement in plasma was significantly higher than that of AFP levels for discriminating HCC from CHB patients (AUC 0.720 vs. 0.571, P<0.001).
Figure 4. Receiver operating characteristic curves (ROC) of AFP, CCND2 promoter methylation, and combined measurement for distinguishing HCC from LC and CHB in both PBMCs and plasma. (A) The area under the ROC curves (AUC) of combined measurement in PBMCs was significantly higher than that of AFP levels for discriminating HCC from LC patients (AUC 0.698 vs. 0.540, P<0.001). (B) The AUC of combined measurement in plasma was significantly higher than that of AFP levels for discriminating HCC from LC patients (AUC 0.694 vs. 0.540, P<0.001). (C) The AUC of combined measurement in PBMCs was significantly higher than that of AFP levels for discriminating HCC from CHB patients (AUC 0.724 vs. 0.571, P<0.001). (D) The AUC of combined measurement in plasma was significantly higher than that of AFP levels for discriminating HCC from CHB patients (AUC 0.720 vs. 0.571, P<0.001). Figure 5. Classification of 118 HCC and 47 LC patients according to AFP concentrations and CCND2 methylation status in plasma. AFP – alpha-fetoprotein; HCC – hepatocellular carcinoma; LC – liver cirrhosis.
Figure 5. Classification of 118 HCC and 47 LC patients according to AFP concentrations and CCND2 methylation status in plasma. AFP – alpha-fetoprotein; HCC – hepatocellular carcinoma; LC – liver cirrhosis. Tables
 Table 1. Characterization of the study participants.
Table 1. Characterization of the study participants. Table 2. Association between CCND2 methylation status and clinical characteristics of HCC patients.
Table 2. Association between CCND2 methylation status and clinical characteristics of HCC patients. Table 3. CCND2 methylation levels in PBMCs and TNM stage from HCC patients.
Table 3. CCND2 methylation levels in PBMCs and TNM stage from HCC patients. Table 4. Diagnostic utility of AFP, CCND2 promoter methylation, and combined measurement for discrimination of HCC from LC and CHB.
Table 4. Diagnostic utility of AFP, CCND2 promoter methylation, and combined measurement for discrimination of HCC from LC and CHB. Table 1. Characterization of the study participants.
Table 1. Characterization of the study participants. Table 2. Association between CCND2 methylation status and clinical characteristics of HCC patients.
Table 2. Association between CCND2 methylation status and clinical characteristics of HCC patients. Table 3. CCND2 methylation levels in PBMCs and TNM stage from HCC patients.
Table 3. CCND2 methylation levels in PBMCs and TNM stage from HCC patients. Table 4. Diagnostic utility of AFP, CCND2 promoter methylation, and combined measurement for discrimination of HCC from LC and CHB.
Table 4. Diagnostic utility of AFP, CCND2 promoter methylation, and combined measurement for discrimination of HCC from LC and CHB. In Press
15 Apr 2024 : Laboratory Research
The Role of Copper-Induced M2 Macrophage Polarization in Protecting Cartilage Matrix in OsteoarthritisMed Sci Monit In Press; DOI: 10.12659/MSM.943738
07 Mar 2024 : Clinical Research
Knowledge of and Attitudes Toward Clinical Trials: A Questionnaire-Based Study of 179 Male Third- and Fourt...Med Sci Monit In Press; DOI: 10.12659/MSM.943468
08 Mar 2024 : Animal Research
Modification of Experimental Model of Necrotizing Enterocolitis (NEC) in Rat Pups by Single Exposure to Hyp...Med Sci Monit In Press; DOI: 10.12659/MSM.943443
18 Apr 2024 : Clinical Research
Comparative Analysis of Open and Closed Sphincterotomy for the Treatment of Chronic Anal Fissure: Safety an...Med Sci Monit In Press; DOI: 10.12659/MSM.944127
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952








