06 March 2024: Clinical Research
Molecular Determinants of Drug Resistance and Mutation Patterns in Influenza Viruses Circulating in Poland Across Multiple Epidemic Seasons: Implications for Vaccination Strategies
Ewelina HallmannDOI: 10.12659/MSM.942125
Med Sci Monit 2024; 30:e942125
Abstract
BACKGROUND: According to the WHO, up to 650 000 people die each year from seasonal flu-related respiratory illnesses. The most effective method of fighting the virus is seasonal vaccination. However, if an infection does occur, antiviral medications should be used as soon as possible. No studies of drug resistance in influenza viruses circulating in Poland have been systematically conducted. Therefore, the aim of the present study was to investigate the drug resistance and genetic diversity of influenza virus strains circulating in Poland by determining the presence of mutations in the neuraminidase gene.
MATERIAL AND METHODS: A total of 258 clinical specimens were collected during the 2016-2017, 2017-2018, and 2018-2019 epidemic seasons. The samples containing influenza A and B were analyzed by RT-PCR and Sanger sequencing.
RESULTS: Differences were found between the influenza virus strains detected in different epidemic seasons, demonstrating the occurrence of mutations. Influenza A virus was found to be more genetically variable than influenza B virus (P<0.001, Kruskal-Wallis test). However, there was no significant difference in the resistance prevalence between the influenza A subtypes A/H1N1/pdm09 (4.8%) and A/H3N2/ (6.1%). In contrast, more mutations of drug-resistance genes were found in the influenza B virus (P<0.001, chi-square test). In addition, resistance mutations appeared en masse in vaccine strains circulating in unvaccinated populations.
CONCLUSIONS: It seems important to determine whether the influenza virus strains tested for drug resistance as part of global influenza surveillance are equally representative of viruses circulating in populations with high and low vaccination rates, for all countries. Our results suggest that countries with low levels of influenza immunization may constitute reservoirs of drug-resistant influenza viruses.
Keywords: Sequence Analysis, RNA, Influenza A virus, Influenza B virus, Poland
Background
The evolution of the influenza virus is most pronounced in the case of surface glycoproteins, but it also affects each of the segments. The accumulation of molecular changes is possible by means of different mechanisms: (a) point mutation (antigenic drift); (b) genetic reassortment (antigenic shift); (c) defective-interfering particles; and (d) RNA recombination [1,2]. These changes may take place alongside other antigenic drift or antigenic shift changes [3]. Mixing of segments from different strains (antigenic shift) gives rise to the possibility of 256 different genotypes [4]. Viral copies that are resistant to one or more drugs – such as neuraminidase inhibitors used to treat or prevent influenza – will be selected for replication in populations using these drugs [5]. Anti-influenza medication should be given as early as possible, preferably within 48 hours from the onset of symptoms. In addition to increasing the drug’s effectiveness, this also reduces the emergence of resistant viruses [6]. There are 3 levels of detecting viral drug resistance: genotypic – detected by sequencing the viral genome and identifying mutants associated with drug resistance; phenotypic – viral drug resistance is determined by measuring resistance at different drug concentrations in vitro; and clinical – based on measuring the response to a drug administered to humans and animals [7]. In Poland, oseltamivir is commercially available for the treatment and prevention of influenza. Its advantage is that it can be used in the full spectrum of age and risk groups [8], but also has a higher risk of developing resistance [9]. Prolonged exposure to oseltamivir during sustained viral replication, suboptimal doses (eg, once-daily prophylactic dosing in people with active infection), and treatment interruptions are reported as risk factors for the development of resistance [10]. By inhibiting neuraminidase activity, oseltamivir prevents the spread of influenza virus from infected cells to healthy ones. However, if the amino acid sequence of influenza neuraminidase proteins is altered, oseltamivir may lose its ability to bind to neuraminidase and inhibit the function of neuraminidase proteins, causing resistance to neuraminidase inhibitors [11,12].
Resistance to inhibitors of neuraminidase is most often associated with a single substitution in a neuraminidase gene. In the influenza A virus, common mutations include H275T in N1 and E119I/V, R292K, N294S, and E119V + I222V in N2 [13,14]. In the influenza B virus, common mutations include D198N/E and R152K [15]. These substitutions prevent oseltamivir from inhibiting neuraminidase activity and cause the mutant virus to spread to healthy cells in patients taking oseltamivir. Both hemagglutinin and neuraminidase are susceptible to neuraminidase inhibitors. Research using chemiluminescence shows that influenza A is more sensitive to drugs than influenza B. Overall resistance to antiviral drugs in viruses isolated worldwide between 2004 and 2008 was low (0.2%). However, in the 2006–2007 epidemic season in the United States, resistance to oseltamivir A/H1N1/ was reported at 0.7%, while in the following season, it was already 10 times higher, at 7.1% [16]. In Norway, in the 2007–2008 epidemic season, resistance due to the H274Y mutation in A/H1N1/ was 67% [17], while in Japan, in the 2008–2009 epidemic season, resistance due to this same mutation was almost 100% [18]. Global influenza surveillance estimated drug resistance to oseltamivir to be <1.5% in the 2009–2010 influenza season [19]. In 2011, in Singapore and Australia, the number of A/H1N1/pdm09 strains resistant to oseltamivir and zanamivir, with 2 mutations, H274Y and S246N, increased by 10% and 30%, respectively [20]. Drug resistance to oseltamivir raises concerns about the emergence of the H274Y mutant among the highly pathogenic avian influenza virus strains; therefore, it is important to monitor drug resistance among influenza viruses in each country. The aim of the present study was to investigate the drug resistance in influenza virus strains circulating in Poland, and to demonstrate their diversity, by determining the occurrence of mutations in the neuraminidase gene.
Material and Methods
CLINICAL SAMPLES:
The research included samples from 3 influenza epidemic seasons (2016–2017, 2017–2018, and 2018–2019), obtained in Poland by the National Influenza Center at the National Institute of Public Health NIH – NRI. Clinical samples collected as part of the global virological and epidemiological surveillance of influenza in Poland (GISRS) were used for this study. Samples included nasal and throat swabs from patients with clinical signs of influenza-like illness, defined as those with a sudden onset of illness, fever >38°C, and cough or sore throat. Samples from patients aged from 1 to 89 years were examined. A total of 258 clinical specimens were analyzed: 91 samples from patients infected with the A/H1N1/pdm09 virus; 105 samples from patients infected with the A/H3N2/ virus; and 62 samples from patients infected with influenza B virus. The samples were stored at −80ºC until the time of testing.
NUCLEIC ACID EXTRACTION OF VIRAL GENETIC MATERIAL:
The viral RNA was isolated using the Qiagen QIAamp Viral RNA Mini Kit (Hilden, Germany). Each time, according to the manufacturer’s instructions, a 50 μL solution containing RNA was obtained from a 140 μL solution of dissolved clinical material [21].
REAL-TIME RT-PCR FOR INFLUENZA TYPE/SUBTYPE DETERMINATION:
A quantitative real-time RT-PCR (qRT-PCR) analysis was performed on the purified RNA extracted from the collected clinical specimens to determine the type/subtype or lineage of influenza virus. The reactions were carried out in capillaries using a Roche Light Cycler 2.0 thermal cycler. The positive control was RNA isolated from the reference viruses used for the vaccine in the analyzed epidemic season, as recommended by the WHO. RNase-free water was used as the negative control. Primer and probe sequences were those recommended by the WHO [22]. Positive influenza samples with CT≤35 after subtyping were then sequenced [23].
CONVENTIONAL RT-PCR TO DETECT MUTATIONS IN THE NEURAMINIDASE GENE:
To detect mutations in the neuraminidase gene, samples were analyzed by means of RT-PCR. Reactions were performed in tubes using a BioRad Thermal Cycler C1000, with a Qiagen OneStep RT-PCR Kit. The reagents were: 10 μL of 5X RT-PCR buffer, 0.6 μM of each of the primers, 400 μM of dNTPs, 2 μL of enzyme mix, 100 ng of template, and RNAse-free water to make a total volume of 50 μL (according to the manufacturer’s instructions). The primers were designed specifically for each influenza A and influenza B subtype [24,25]. The reaction conditions were established experimentally, taking into account the melting points of the individual primers (Table 1).
Amplification involved an initial reverse transcription step at 50°C for 30 min followed by initial denaturation at 95°C for 15 min, 35 cycles of: denaturation at 94°C for 1 min; annealing for 1 min at 63°C for A/H1N1/pdm09, at 58°C for A/H3N2/, and at 56°C for influenza type B; extension at 72°C for 1 min; and a final extension step at 72°C for 10 min.
GEL ELECTROPHORESIS AND ISOLATION OF PCR PRODUCTS:
The electrophoretic separation of the PCR products obtained and their isolation from the gel allowed a higher concentration of DNA and its desalting, which has a positive effect on the quality of the sequencing reaction [26]. Agarose gel with a concentration of 1.9% was used with Midori Green Advance nucleic acid dye [27]. Electrophoresis was carried out for 90 minutes at 80 volts. The extraction of PCR products from the gel was carried out on columns according to the manufacturer’s instructions (A&A Biotechnology, Gdańsk, Poland). DNA concentration was checked using the NanoDrop One apparatus (ThermoFisher, Waltham, USA).
SANGER SEQUENCING OF SAMPLES WITH DETECTED MUTATIONS:
Sequencing was carried out using a DTCS Quick Start Kit (Beckman Coulter, Brea, USA), according to the manufacturer’s instructions, with a C1000 BioRad Thermal Cycler, in the laboratory of the Virology Department in the National Institute of Public Health NIH-NRI. The analysis was performed using the GenomeLab GeXP by Beckman Coulter [28,29]. Each type of influenza virus was analyzed in a separate 96-well plate to minimize the possibility of sample contamination [30].
SEQUENCE ANALYSIS:
The analysis of nucleotide sequences was carried out using the following software: MEGA 6 [31], CLC Main Workbench. The resulting protein sequences were then analyzed using BioEdit version 7.0.5.3 [32] and Clustal W [33] to identify resistance-associated mutations, also using the FluSurver bioinformatics tool. The FluSurver application was also used to create figures representing mutations.
STATISTICAL ANALYSIS:
Descriptive statistics methods, including box-plots and statistical tests, were used in the results analysis. The statistical significance of differences between the tested virus strains in terms of the prevalence of detected mutations, including those conferring resistance, was assessed using the chi-square test. The same test was used to compare mutation rates in 2 age groups: ≤14 years of age and ≥15 years of age. The Mann-Whitney test was used to compare the distribution of the number of mutations between 2 groups (based on virus subtypes and age categories,), while the Kruskal-Wallis test was used when larger numbers of groups were compared. (In this case, after showing statistically significant differences between 2 groups, to identify different pairs, Dunn’s post-hoc test was used.)
A significance level of 0.05 was used for all the statistical tests. The analyses were performed using SPSS 12.0 PL statistical software.
Results
REAL-TIME RT-PCR:
Influenza A virus was found in 91 samples, which contained the A/H1N1/pdm09 virus genetic material, and 105 samples, which contained the A/H3N2/ virus genetic material. Influenza B virus was found in 62 samples, of which 91.3% were the Yamagata lineage of influenza B virus.
SANGER SEQUENCING OF SAMPLES:
The influenza viruses were sequenced. Results were obtained from 193 clinical specimens: 36 samples were from patients infected with the A/H1N1/pdm09 virus; 100 samples were from patients infected with the A/H3N2/ virus; and 57 samples were from patients infected with the influenza B virus.
DESCRIPTION OF DETECTED MUTATIONS:
The percentage of influenza A sequences with mutations in the neuraminidase gene was high: 86.1% (31/36) for A/H1N1/pdm09, 89% (89/100) for A/H3N, 71.9% (41/57) for influenza B (Table 2). Regarding mutations conferring drug resistance, they were found in 50% (18/36), 22% (22/100) and 47.4% (27/57) of samples of influenza A/H1N1/pdm09, influenza A/H3N2, and influenza B, respectively. Mutations conferring drug resistance among influenza viruses were detected in 35% of all the clinical specimens.
STATISTICAL ANALYSIS OF MUTATIONS:
No statistically significant differences in resistance prevalence were detected between subtypes A/H1N1/pdm09 (5.4%) and A/H3N2/ (6.3%) (chi-square test). In the case of the influenza B virus, the frequency of drug-resistant mutations among the overall detected mutations was significantly higher (19.1%) than in the influenza A virus samples (5.8%) (
TYPES/PROPERTIES OF OTHER MUTATIONS DETECTED:
In addition to drug-resistance mutations, other types of mutations were also detected in the influenza A and B viruses, as shown in Table 3.
Discussion
Resistance to anti-influenza drugs in Europe is seen at different levels. The highest level of neuraminidase inhibitor resistance in the 2008 A/H1N1/ virus strain was found in Norway, where resistance was seen in 70% of the viruses, while in France the percentage was 17%. By contrast, resistance was detected in less than 10% of the viruses in the UK, the Netherlands, and Germany. In turn, neither Austria nor France reported any drug-resistant strains at that time [35]. Drug resistance prevalence in Poland in the 3 analyzed epidemic seasons, taking into account all the samples analyzed, was 33%. Anti-influenza drugs – neuraminidase inhibitors – do not replace vaccination, but these constitute an effective method for preventing post-influenza complications. Neuraminidase inhibitors also prove useful when a new strain of the influenza virus appears in the population that is significantly different from the viruses that were used to make the vaccine for a given epidemic season. Neuraminidase inhibitors play an essential role as these target the highly conserved neuraminidase active site [36].
Studies of viruses from the 2016–2017, 2017–2018, and 2018–2019 seasons showed that almost all of the detected infections with drug-resistant strains could be prevented by vaccination. Among the tested samples with detected mutations associated with drug resistance, both the subtype A/H1N1/pdm09 and the influenza B viruses were vaccine strains. The presented data demonstrate the importance of influenza vaccination in terms of limiting the spread of infections and the emergence of drug-resistant strains. This also confirms the accuracy of strain selection for vaccine development in the seasons studied.
Prior to the introduction of neuraminidase inhibitors in 1999, drug resistance rates were estimated to be <1% [37]. The first reports of influenza viruses resistant to neuraminidase inhibitors come from the 2007–2008 epidemic season [38]. When WHO influenza expert Frederick Hayden announced that a single oseltamivir-resistant mutation (H275Y substitution in neuraminidase) had been discovered in Europe, it caused a wave of concern. One of the best tools to fight the flu might no longer be effective. In Japan in the 2007–2008 epidemic season, resistant viruses belonging to clade 2B A/Brisbane/59/2007 were detected; these viruses had originated from the European cluster [39]. Therefore, it can be concluded that Europe was the cradle of this mutant variant. It has also been concluded that resistance is largely due to the spread of a single variant [40]. Now, more than 10 years after this discovery, knowledge about not only the phenomenon of drug resistance, but also new anti-influenza drugs, vaccinations, or the genetics of the influenza virus itself, has significantly increased through the work of scientists and medical doctors [41,42]. Assessment of the susceptibility of currently circulating influenza viruses to antivirals is an important element of influenza virological surveillance required by the WHO, which the National Influenza Centers are trying to fulfill [43]. The WHO regularly updates the list of detected mutations [44]. As part of influenza surveillance, susceptibility to neuraminidase inhibitors is routinely assessed phenotypically, using a neuraminidase inhibition assay on selected samples. PA cap-dependent endonuclease, neuraminidase, and M2 gene sequence analysis is used to identify known markers of molecular resistance to all 3 classes of antiviral drugs [45]. However, it is important for scientists to also have information about other types of mutations that may be helpful in designing new drugs. A way to alleviate the problem of drug resistance is to design drugs that target core structural components or enzymatic active sites of viral proteins, because mutations in these residues likely produce structurally unstable or inactive proteins. In the future, the main way to combat influenza infection may be to select combinations of drugs with different mechanisms of action in response to drug failure due to viral mutations [46]. In the present study, we detected different types of mutations, which are described in the results section. The highest percentage of mutations detected in all types of influenza viruses occurred in genes related to ligand-binding and oligomerization properties of the virus. The viral oligomerization interface is an area responsible for the stability of neuraminidase oligomerization. A better understanding of the biological functions of neuraminidase may facilitate an alternative design for antiviral drugs to combat influenza virus infection. Notably, escape mutations occurred 644 times worldwide (6.89% prevalence). In the present study, such mutations were found 3 times in the influenza B virus and twice in the influenza A virus. They evade the host’s immune system response by no longer being effectively recognized by neutralizing antibodies. This is possible, for example, by producing antigen proteins that are similar to host proteins [47]. Based on published information, N-linked glycosylation may be important in adhesion with host cells or the surrounding environment, and may play a role in antigenicity [48]. Recent studies have shown that many viral proteins, especially structural proteins, are glycosylated during the viral infection cycle. The N-glycans of the viral glycoproteins have multiple functions, which include promotion of expression, transport, fusion, binding to cell surface receptors, and prevention of antibody neutralization [49,50].
In Brazil, studies were conducted in 2017–2019 that confirmed the presence of adamantane resistance markers M2: S31N in most of the viruses tested, in line with what has been reported around the world. No increased resistance of viruses to neuraminidase inhibitors has been reported. Despite this, this situation can change rapidly, and resistant-strain surveillance is a priority. This highlights the importance of neuraminidase inhibitors in the treatment of influenza infections [51]. During the COVID-19 pandemic, measures taken to limit the spread of SARS-CoV-2 led to a substantial decrease in global influenza activity in the 2020–21 and 2021–22 epidemic seasons [52]. Data from WHO influenza surveillance in 2022 revealed laboratory-confirmed cases of influenza well above the historical average [53]. Analysis of drug-resistant influenza viruses carried out in Germany in 2019–2022 showed that resistance to influenza antiviral drugs may develop in the absence of selection pressure, spontaneously even, in untreated patients. Circulation and wider spread of such viral quasi-species, carrying smaller, resistant gene variants, can lead to the failure of an antiviral drug or even an entire class of drugs [54]. In Poland, the National Influenza Center has not yet performed studies from the post-pandemic period. The number of samples collected in the country in the 2020–2021 and 2021–2022 seasons did not allow for detailed analyses. The CDC reports that antiviral susceptibility patterns changed very little during the 2020–2021 and 2021–2022 seasons. Only a very small number of viruses were found to be resistant to oseltamivir [55].
The use of anti-influenza drugs for treatment depends on the policy of a given country. The available neuraminidase inhibitor in Poland is oseltamivir [56]. Japan has been one of the largest consumers of neuraminidase inhibitors in the world [57]. However, in recent years, the statistics have changed, and now the United States is ahead of Japan in neuraminidase inhibitor use. According to media information provided by F. Hoffmann-La Roche, in the period between January and December 2014, the United States accounted for 71.5% of the global consumption of neuraminidase inhibitors, Japan accounted for 11.8%, Europe accounted for 7.7%, and the rest of the world accounted for 9% [58]. Due to the percentage of the adult population that is vaccinated in Japan, which was 19.17% in 2011; 17.17% in 2012; and 42.86% in patients with immunodeficiency, it can be presumed that the high use of anti-influenza vaccines in the local population did not affect the development of resistance among viruses, because this phenomenon should be eliminated by vaccination. A similar situation also applies to the United States, as mentioned previously, where 36.2% of the population has been vaccinated against influenza [59]. In Poland, a very small part of the population gets vaccinated against influenza – in the analyzed epidemic seasons, vaccination in the population ranged from 3.3 to 3.9% [60,61]. In the previously discussed countries, in the corresponding epidemic seasons, the percentage of the population that was vaccinated was on average 42.2% in the USA [62], and similarly in Japan, about 40% [63].
Contrary to what was observed in Japan [64], we observed a higher frequency of mutations within the neuraminidase gene in the influenza A strains in Poland. Moreover, unlike the observations from Japan, all the strains detected in the present study were vaccine strains. These data suggest that the observed differences in drug resistance mutation incidence in influenza A and B viruses may depend on the level of vaccination of the human population from which the tested influenza virus isolates were obtained. Therefore, it seems important to determine whether the strains of influenza viruses tested for drug resistance as part of global influenza surveillance are representative of high and low vaccination rates of the population to a similar extent for all countries. Our results suggest that countries with low levels of influenza immunization may constitute reservoirs of drug-resistant influenza viruses. Drug resistance studies of influenza virus strains in other countries with a low level of vaccination in their population should be conducted to assess the scale of the risk of new drug-resistant strains arising from the lack of influenza vaccination.
An interesting study was conducted in the United States which estimated that for every 100 children under the age of 15 hospitalized for cardiopulmonary diseases during the flu season, 3 to 9 courses of antibiotic therapy were prescribed each year. The study encompassed 19 years of observations using the collected data and calculated the incidence of influenza in each given period. It turned out that 10–30% of excessive use of antibiotics during the winter period may be due to influenza [65]. A meta-analysis conducted in 2019 showed that influenza vaccination reduced the duration of antibiotic therapy among healthy adults by 28.1% and moderately in children from 6 months to 14 years of age [66]. This suggests that the use of prophylaxis in the form of vaccination works synergistically with treatment, and taking advantage of this synergy may be an approach for reducing the development of antiviral resistance. Vaccination can reduce the evolution of resistant variants, while drugs can subsequently be used against strains for which vaccination has not provided adequate protection. The broad use of neuraminidase inhibitors, coupled with a low level of vaccination of the population, may serve to create selection pressure driving the evolution of resistant strains. The present study is the first of this type conducted in Poland. It would be helpful to extend this research by employing phenotype-based drug resistance tests and comparing the results with the mutations detected using the genotypic method.
Conclusions
All viruses in which mutations were detected were the same strains as those in the seasonal influenza vaccine in use during the examined epidemic season.
Figures
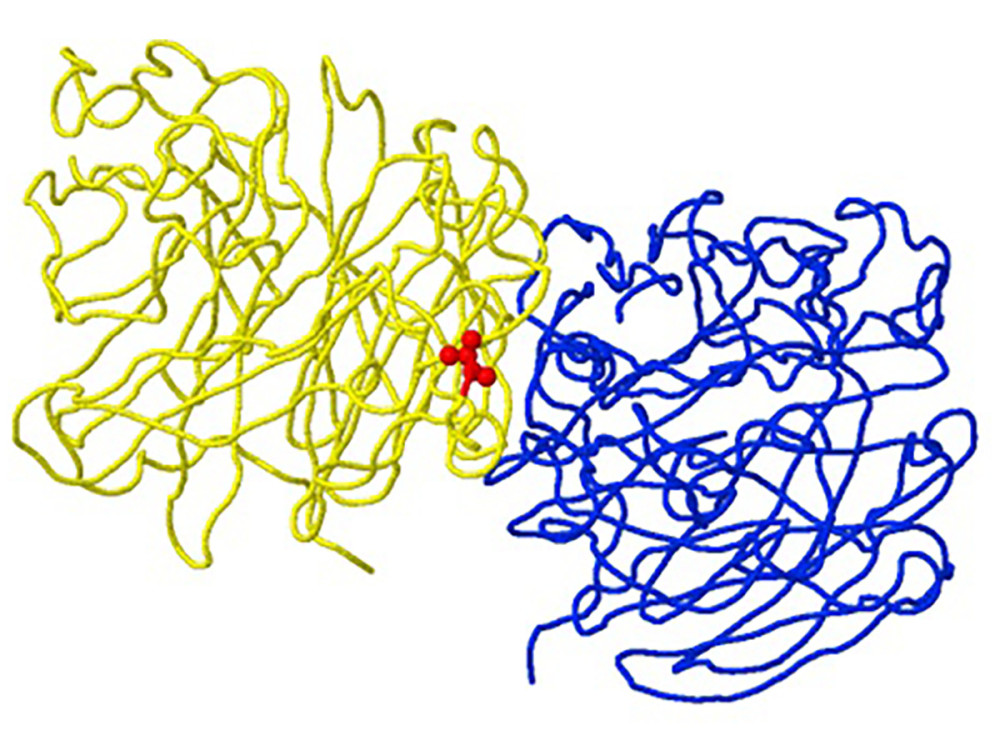 Figure 1. Structure of neuraminidase A/Brisbane/02/2018 (A/H1N1/pdm09) with the R77G mutation detected in clinical sample number 145, as an example of a “viral oligomerization interface” mutation. The location of the mutation corresponds to position 77 of the B subunit chain (yellow backbone) and is 5 Ångström (A) from the A subunit oligomeric chain (blue backbone) in the A/H1N1/pdm09 neuraminidase gene (created using the FluSurver mutations app – https://gisaid.org/database-features/flusurver-mutations-app/).
Figure 1. Structure of neuraminidase A/Brisbane/02/2018 (A/H1N1/pdm09) with the R77G mutation detected in clinical sample number 145, as an example of a “viral oligomerization interface” mutation. The location of the mutation corresponds to position 77 of the B subunit chain (yellow backbone) and is 5 Ångström (A) from the A subunit oligomeric chain (blue backbone) in the A/H1N1/pdm09 neuraminidase gene (created using the FluSurver mutations app – https://gisaid.org/database-features/flusurver-mutations-app/). 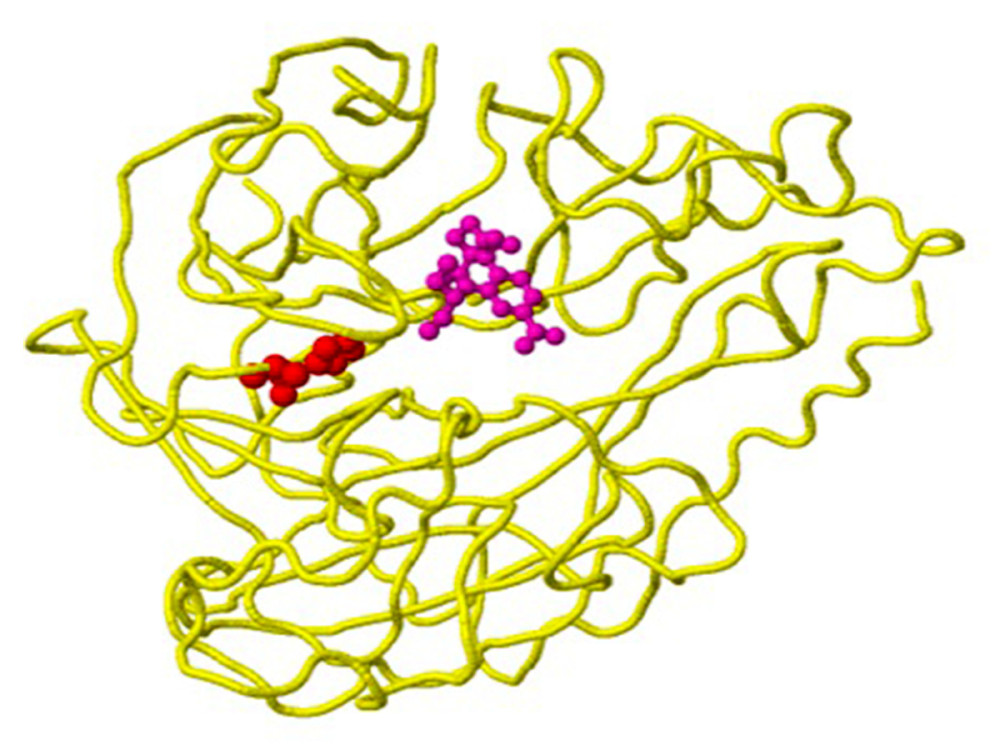 Figure 2. Structure of the neuraminidase of the virus B/Phuket/3073/2013 with the D356H mutation detected in clinical sample number 203, as an example of a mutation with the property of drug binding. The location of the mutation corresponds to position 224 of the A chain (yellow backbone) and is 5 Ångström (A) from the drug-binding site (created using the FluSurver mutations app – https://gisaid.org/database-features/flusurver-mutations-app/).
Figure 2. Structure of the neuraminidase of the virus B/Phuket/3073/2013 with the D356H mutation detected in clinical sample number 203, as an example of a mutation with the property of drug binding. The location of the mutation corresponds to position 224 of the A chain (yellow backbone) and is 5 Ångström (A) from the drug-binding site (created using the FluSurver mutations app – https://gisaid.org/database-features/flusurver-mutations-app/). 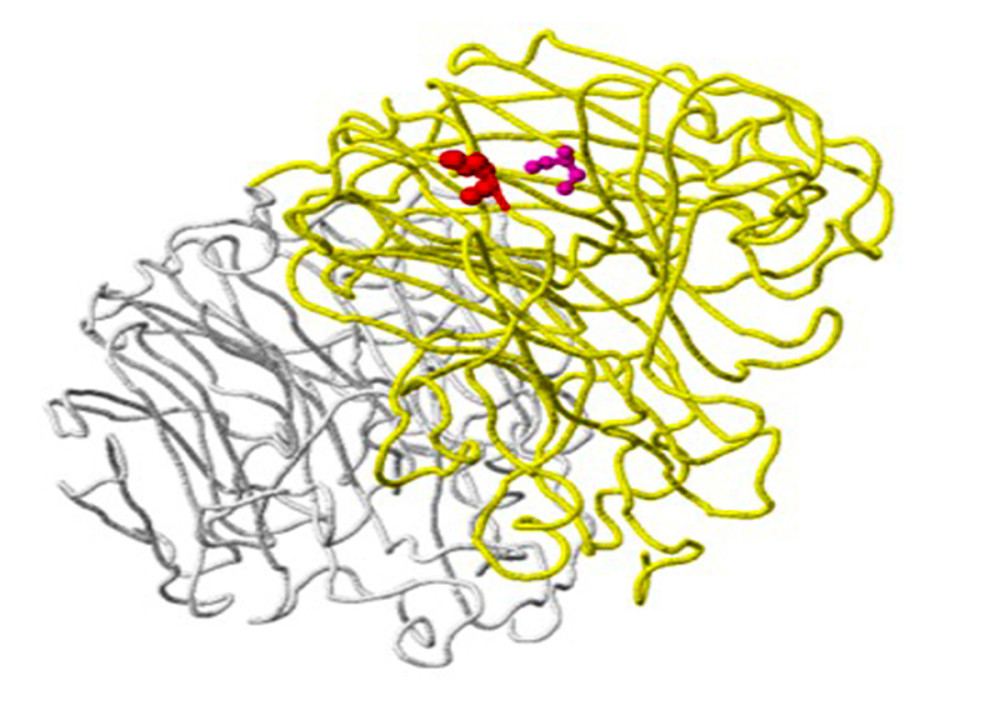 Figure 3. Structure of the neuraminidase virus A/SingaporeINFIMH-16-0019/2016(A/H3N2/) with the D356H mutation detected in clinical sample number 25, as an example of a ligand-binding mutation. Mutation site 356 is shown in red on the A chain (yellow backbone), which is 5 Ångström (A) from the ligand-binding site (created using the FluSurver mutations app – https://gisaid.org/database-features/flusurver-mutations-app/).
Figure 3. Structure of the neuraminidase virus A/SingaporeINFIMH-16-0019/2016(A/H3N2/) with the D356H mutation detected in clinical sample number 25, as an example of a ligand-binding mutation. Mutation site 356 is shown in red on the A chain (yellow backbone), which is 5 Ångström (A) from the ligand-binding site (created using the FluSurver mutations app – https://gisaid.org/database-features/flusurver-mutations-app/). 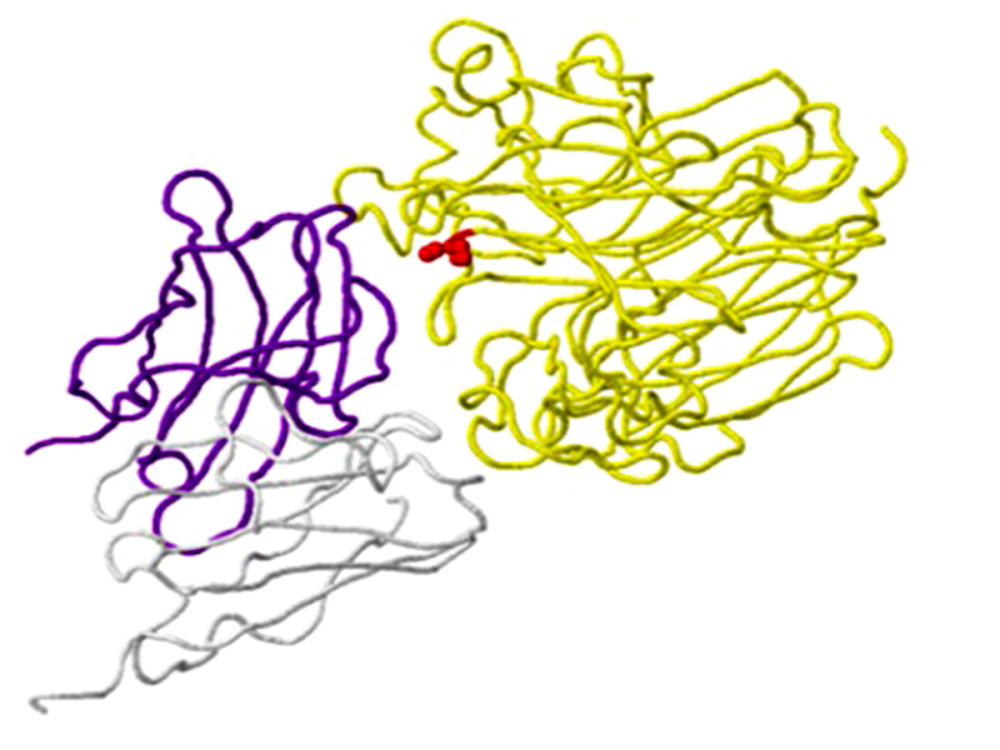 Figure 4. An example of a mutation site related to antibody-binding properties: the K369N mutation in the neuraminidase gene of virus A/Brisbane/02/2018 (A/H1N1/pdm09) in clinical sample number 770. The location of the mutation (red) corresponds to position 369 on the viral N chain (yellow backbone) and is 5 Ångström (A) from the heavy chain (H) antibodies (purple skeleton) (created using the FluSurver mutations app – https://gisaid.org/database-features/flusurver-mutations-app/).
Figure 4. An example of a mutation site related to antibody-binding properties: the K369N mutation in the neuraminidase gene of virus A/Brisbane/02/2018 (A/H1N1/pdm09) in clinical sample number 770. The location of the mutation (red) corresponds to position 369 on the viral N chain (yellow backbone) and is 5 Ångström (A) from the heavy chain (H) antibodies (purple skeleton) (created using the FluSurver mutations app – https://gisaid.org/database-features/flusurver-mutations-app/). 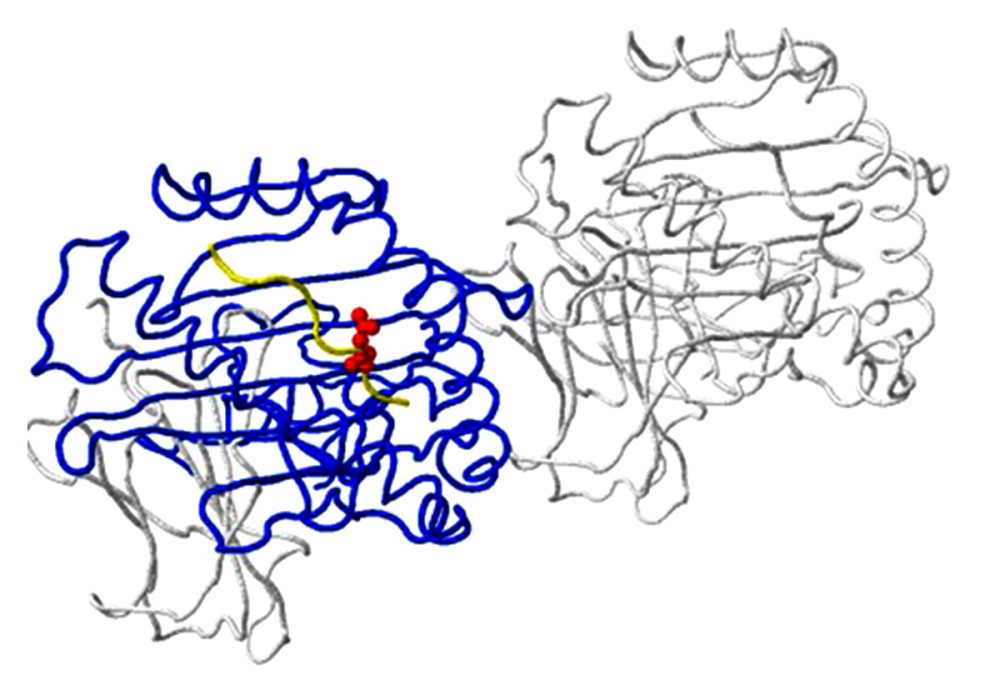 Figure 5. Host protein-binding site exemplified by the G451R mutation in the neuraminidase gene of virus A/SingaporeINFIMH-16-0019/2016 (A/H3N2/) in clinical sample number 229. The location of the mutation corresponds to position 452 of the viral F chain (yellow backbone) of the neuraminidase gene and is 5 Ångström (A) from the D-chain of a non-viral protein (blue backbone), in this case the MHC class I antigen. The white backbone represents the other chains found in the structure (created using the FluSurver mutations app – https://gisaid.org/database-features/flusurver-mutations-app/).
Figure 5. Host protein-binding site exemplified by the G451R mutation in the neuraminidase gene of virus A/SingaporeINFIMH-16-0019/2016 (A/H3N2/) in clinical sample number 229. The location of the mutation corresponds to position 452 of the viral F chain (yellow backbone) of the neuraminidase gene and is 5 Ångström (A) from the D-chain of a non-viral protein (blue backbone), in this case the MHC class I antigen. The white backbone represents the other chains found in the structure (created using the FluSurver mutations app – https://gisaid.org/database-features/flusurver-mutations-app/). 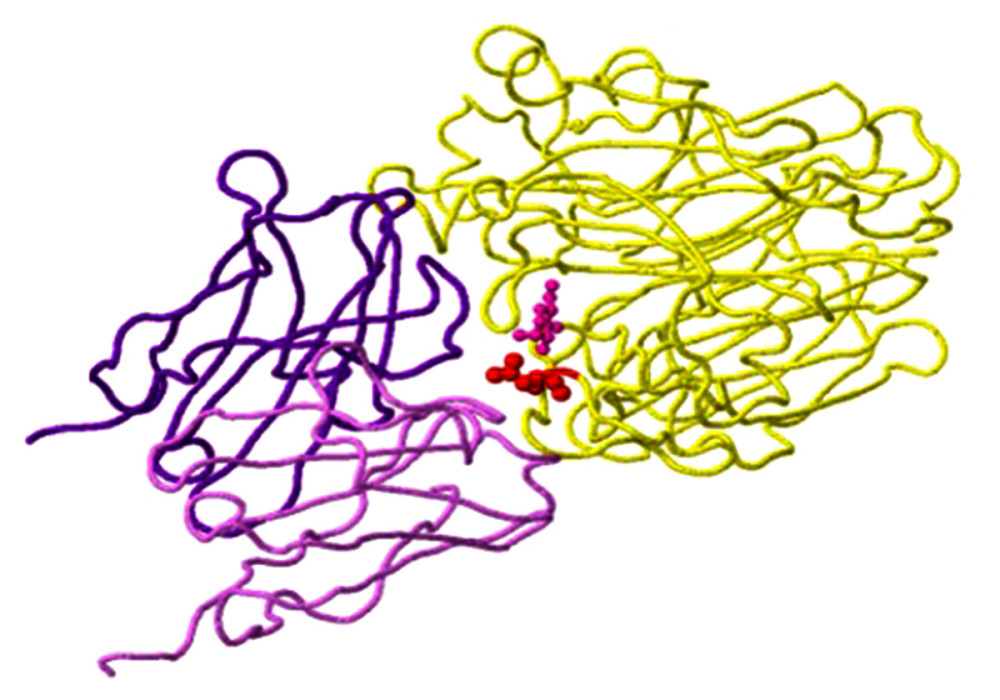 Figure 6. N-glycosylation on the example of the detected N329S mutation in the neuraminidase gene of virus A/SingaporeINFIMH-16-0019//2016 (A/H3N2/) in clinical sample number 30598. The mutation site corresponds to position 329 of the viral N chain (yellow backbone) and is located 5 Ångströms (A) from the ligand, the antibody heavy (H) chain (purple backbone), and the antibody light (L) chain (light purple backbone) (created using the FluSurver mutations app – https://gisaid.org/database-features/flusurver-mutations-app/).
Figure 6. N-glycosylation on the example of the detected N329S mutation in the neuraminidase gene of virus A/SingaporeINFIMH-16-0019//2016 (A/H3N2/) in clinical sample number 30598. The mutation site corresponds to position 329 of the viral N chain (yellow backbone) and is located 5 Ångströms (A) from the ligand, the antibody heavy (H) chain (purple backbone), and the antibody light (L) chain (light purple backbone) (created using the FluSurver mutations app – https://gisaid.org/database-features/flusurver-mutations-app/). Tables
Table 1. Sequences of primers used in the RT-PCR for the detection of selected fragments of genes associated with resistance to anti-influenza drugs.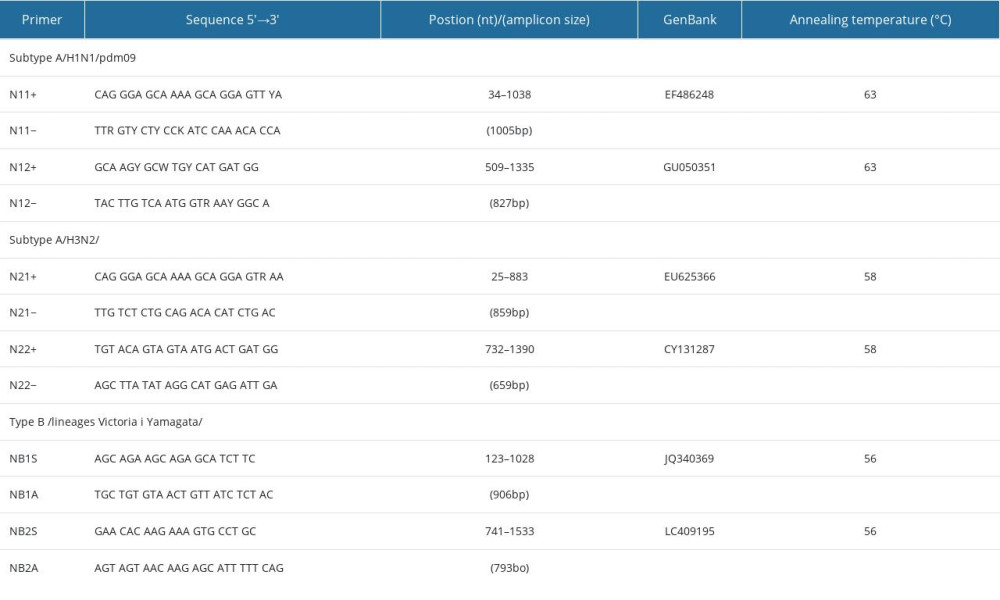 Table 2. Summary of results related to the detected mutations in the influenza viruses tested.
Table 2. Summary of results related to the detected mutations in the influenza viruses tested.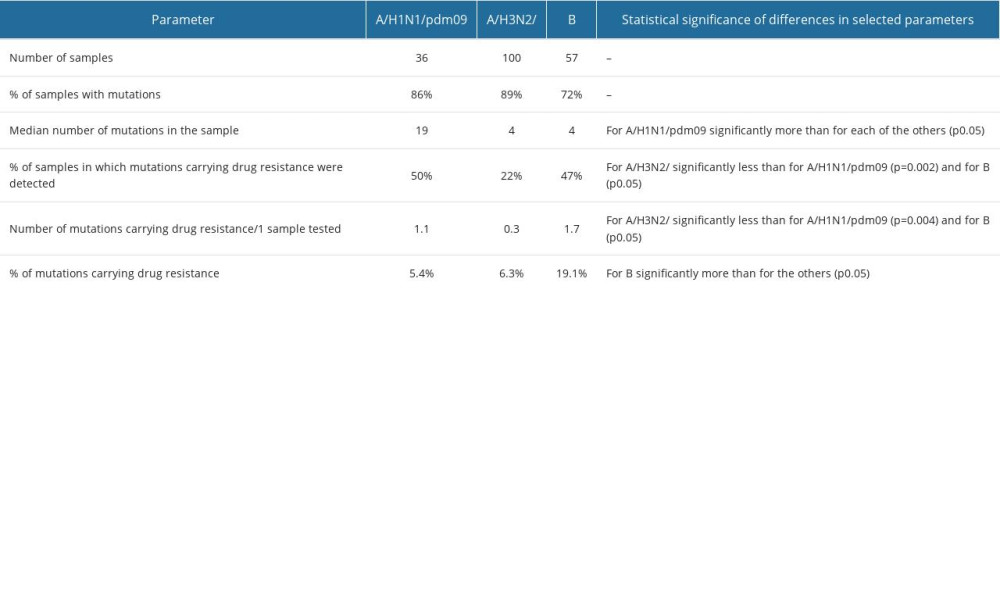 Table 3. Percentage distribution of other properties at the site of detected mutations in influenza viruses in Poland.
Table 3. Percentage distribution of other properties at the site of detected mutations in influenza viruses in Poland.
References
1. Nicholson KG, Wood JM, Zambon M, Influenza: Lancet, 2003; 362; 1733-45
2. Scholtissek C, Molecular evolution of influenza viruses: Virus Genes, 1996; 11; 209-15
3. Brydak LB, Flu. Flu pandemic: Myth or real threat, 2008; 253-82, Warsaw, RYTM Publishing House
4. Taubenberger JK, Kash JC, Influenza virus evolution, host adaptation, and pandemic formation: Cell Host Microbe, 2010; 7(6); 440-51
5. Centers for Disease Control and Prevention (CDC): Influenza Antiviral Drug Resistance. Questions & Answers, 2022 https://www.cdc.gov/flu/treatment/antiviralresistance.htm
6. Ciesla G, Leader S, Stoddard J, Antibiotic prescribing rates in the US ambulatory care setting for patients diagnosed with influenza, 1997–2001: Respir Med, 2004; 98; 1093-101
7. Nitsch-Osuch A, Brydak LB, Influenza viruses resistant to neuraminidase inhibitors: Acta Biochim Pol, 2014; 61(3); 505-8
8. Garg S, Fry AM, Patton M, Antiviral treatment of influenza in children: Pediatr Infect Dis J, 2013; 31(2); e43-51
9. Hurt AC, Chotpitayasunondh T, Cox NJ, Antiviral resistance during the 2009 influenza A H1N1 pandemic: Public health, laboratory, and clinical perspectives: Lancet Infect Dis, 2012; 12; 240-48
10. Li TC, Chan MC, Lee N, Clinical implications of antiviral resistance in influenza: Viruses, 2015; 7(9); 4929-44
11. Webster RG, Monto AS, Braciale TJ, Lamb RA: Textbook of Inluenza. Part 2 Structure and replication, 2013, UK, John Wiley & Sons
12. Hussain M, Galvin HD, Haw TY, Drug resistance in influenza A virus: The epidemiology and management: Infect Drug Resist, 2017; 10; 121-34
13. Okomo-Adhiambo M, Demmler-Harrison GJ, Deyde VM, Detection of E119V and E119I mutations in influenza A (H3N2) viruses isolated from an immunocompromised patient: Challenges in diagnosis of oseltamivir resistance: Antimicrob Agents Chemother, 2010; 54(5); 1834-41
14. Simon P, Holder BP, Bouhy X, The I222V neuraminidase mutation has a compensatory role in replication of an oseltamivir-resistant influenza virus A/H3N2 E119V mutant: J Clin Microbiol, 2011; 49(2); 715-17
15. Nguyen HT, Fry AM, Gubareva LV, Neuraminidase inhibitor resistance in influenza viruses and laboratory testing methods: Antiviral Ther, 2012; 17; 159-73
16. Sheu TG, Deyde VM, Okomo-Adhiambo M, Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide from 2004 to 2008: Antimicrob Agents Chemother, 2008; 52(9); 3284-92
17. Hauge SH, Blix HS, Borgen K, Sales of oseltamivir in Norway prior to the emergence of oseltamivir resistant influenza A(H1N1) viruses in 2007–08: Virol J, 2009; 6; 54
18. Ujike M, Shimabukuro K, Mochizuki K, Oseltamivir-resistant influenza viruses A/H1N1/ during 2007–2009 influenza seasons, Japan: Emerg Infect Dis, 2010; 16; 926-35
19. Ujike M, Ejima M, Anraku A, Monitoring and characterization of Oseltamivir-resistant pandemic A/H1N1/2009 virus, Japan, 2009–2010: Emerg Infect Dis, 2011; 17; 470-79
20. Hurt AC, Lee RT, Leang SK, Increased detection in Australia and Singapore of a novel influenza A(H1N1)2009 variant with reduced oseltamivir and zanamivir sensitivity due to a S247N neuraminidase mutation: Euro Surveill, 2011; 16(23); 19884
21. Bednarska K, Hallmann-Szelińska E, Kondratiuk K, Brydak LB, Evaluation of the activity of influenza and influenza-like viruses in the epidemic season 2013/2014: Adv Exp Med Biol, 2015; 857; 1-7
22. World Health Organization: WHO information for the molecular detection of influenza viruses, 2020 https://cdn.who.int/media/docs/default-source/influenza/global-influenza-surveillance-and-response-system/related-documents/protocols_influenza_virus_detection_jan_2020.pdf?sfvrsn=36349aa3_8
23. Monamele GC, Vernet MA, Njankouo MR, Genetic and antigenic characterization of influenza A(H3N2) in Cameroon during the 2014–2016 influenza seasons: PLoS One, 2017; 12(9); e0184411
24. Chander Y, Jindal N, Stallknecht DE, Full length sequencing of all nine subtypes of the neuraminidase gene of influenza A viruses using subtype specific primer sets: J Virol Methods, 2010; 165(1); 116-20
25. Zhou B, Lin X, Wang W, Universal influenza B virus genomic amplification facilitates sequencing, diagnostics, and reverse genetics: J Clin Microbiol, 2014; 52(5); 1330-37
26. Lee PY, Costumbrado J, Hsu CY, Kim YH, Agarose gel electrophoresis for the separation of DNA Fragments: J Vis Exp, 2012; 62; e3923
27. Hallmann-Szelińska E, Bednarska K, Kondratiuk K, Viral infections in children in the 2014/2015 epidemic season in Poland: Adv Exp Med Biol, 2016; 912; 51-56
28. Sanger F, Nicklen S, Coulson AR, DNA sequencing with chain-terminating inhibitors: Proc Natl Acad Sci USA, 1977; 74; 5463-67
29. Bao JR, Huard TK, Piscitelli AE, Reverse – transcription polymerase chain reaction/pyrosequencing to characterize neuraminidase H275 residue of influenza A 2009 H1N1 virus for rapid and specific detection of the viral oseltamivir resistance marker in a clinical laboratory: Diagn Microbiol Infect Dis, 2011; 71; 396-402
30. Ghedin E, Sengamalay NA, Shumway M, Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution: Nature, 2005; 437; 1162-66
31. Tamura K, Stecher G, Peterson D, MEGA6: molecular evolutionary genetics analysis version 6.0: Mol Biol Evol, 2013; 30; 2725-29
32. Hall TA, BioEdit: A user-friendly biological sequence alignment editor and analysis for Windows 95/98/NT: Nucleic Acids Symp Ser, 1999; 41; 95-98
33. Chenna R, Sugawara H, Koike T, Multiple sequence alignment with the Clustal series of programs: Nucleic Acids Res, 2003; 31; 3497-500
34. Bhat AH, Maity S, Giri K, Ambatipudi K, Protein glycosylation: Sweet or bitter for bacterial pathogens?: Crit Rev Microbiol, 2019; 45; 82-102
35. Meijer A, Lackenby A, Hungnes O, Oseltamivir-resistant influenza virus A (H1N1), Europe, 2007–08 season: Emerg Infect Dis, 2009; 15; 552-60
36. Yen HL, Hoffmann E, Taylor G, Importance of neuraminidase active-site residues to the neuraminidase inhibitor resistance of influenza viruses: J Virol, 2006; 80; 8787-95
37. Hurt AC, Barr IG, Influenza viruses with reduced sensitivity to the neuraminidase inhibitor drugs in untreated young children: Commun Dis Intell, 2008; 32; 57-62
38. Lackenby A, Hungnes O, Dudman SG, Emergence of resistance to oseltamivir among influenza A(H1N1) viruses in Europe: Euro Surveill, 2008; 13; 8026
39. Matsuzaki Y, Mizuta K, Aoki Y, A two-year survey of the oseltamivir-resistant influenza A(H1N1) virus in Yamagata, Japan and the clinical effectiveness of oseltamivir and zanamivir: Virol J, 2010; 7; 53
40. Lampejo T, Influenza and antiviral resistance: An overview: Eur J Clin Microbiol Infect Dis, 2020; 39; 1201-8
41. Smyk JM, Szydłowska N, Szulc W, Majewska A, Evolution of influenza viruses – drug resistance, treatment options, and prospects: Int J Mol Sci, 2022; 23; 12244
42. Stannard HL, Mifsud EJ, Wildum S, Assessing the fitness of a dual-antiviral drug resistant human influenza virus in the ferret model: Commun Biol, 2022; 5(1026)
43. Lackenby A, Besselaar TG, Daniels RS, Global update on the susceptibility of human influenza viruses to neuraminidase inhibitors and status of novel antivirals, 2016–2017: Antiviral Res, 2018; 157; 38-46
44. World Health Organization: Summary of neuraminidase (NA) amino acid substitutions assessed for their effects on inhibition by neuraminidase inhibitors (NAIs), 2023 https://cdn.who.int/media/docs/default-source/global-influenza-programme/1.-nai_human_reduced-susceptibility-marker-table-(who)_07.03.23_update.pdf?sfvrsn=eef56b17_1&download=true
45. Duwe SC, Schmidt B, Gärtner BC, Prophylaxis and treatment of influenza: Options, antiviral susceptibility, and existing recommendations: GMS Infect Dis, 2021; 9; Doc02
46. Li Y, Huo S, Yin Z, The current state of research on influenza antiviral drug development: drugs in clinical trial and licensed drugs: mBio, 2023; 14; e0127323
47. Doud MB, Lee JM, Bloom JD, How single mutations affect viral escape from broad and narrow antibodies to H1 influenza hemagglutinin: Nat Commun, 2018; 9; 1386
48. Zabczynska M, Pochec E, The role of protein glycosylation in immune system: Post Biochem, 2015; 61; 129-37
49. Sealy JE, Peacock TP, Sadeyen JR, Adsorptive mutation and N-linked glycosylation modulate influenza virus antigenicity and fitness: Emerg Microbes Infect, 2020; 9; 2622-31
50. Feng T, Zhang J, Chen Z, Glycosylation of viral proteins: Implication in virus-host interaction and virulence: Virulence, 2022; 13; 670-83
51. Sousa TDC, Martins JSCC, Miranda MD, Low prevalence of influenza A strains with resistance markers in Brazil during 2017–2019 seasons: Front Public Health, 2022; 10; 944277
52. Oh DY, Buda S, Biere B, Trends in respiratory virus circulation following COVID-19-targeted nonpharmaceutical interventions in Germany, January – September 2020: Analysis of national surveillance data: Lancet Reg Health Europe, 2021; 6; 100112
53. World Health Organization, 2022 Available from: https://www.who.int/teams/global-influenza-programme/surveillance-and-monitoring/influenza-updates/influenza-updates-archive
54. Oh DY, Milde J, Ham Y, Preparing for the next influenza season: Monitoring the emergence and spread of antiviral resistance: Infect Drug Resist, 2023; 16; 949-59
55. Centers for Disease Control and Prevention (CDC), 2022 Available from: https://www.cdc.gov/flu/treatment/antiviralresistance.htm
56. : Recommendation for management of respiratory infections, National Medicines Institute Available from:[in Polish]https://antybiotyki.edu.pl/wpcontent/uploads/Rekomendacje/Rekomendacje2016pdf
57. Nguyen-Van-Tam JS, Venkatesan S, Neuraminidase inhibitors: Who, when, where?: Clin Microbiol Infect, 2015; 21; 222-25
58. F. Hoffmann-La Roche Ltd: Media Release. Roche delivers solid results in 2014 Available from:http://www.roche.com/med-cor-2015-01-28-e.pdf
59. Kumar M, Fukuda T, Stankus AP, DiBonaventura M, Influenza vaccination in Japan among the general population and high-risk groups: Value Health, 2014; 17(7); A804
60. Brydak LB, Epidemic season 2019/2020 – what’s new in flu?: Therapy, 2019; 12; 6-10
61. : Epimeld, 2020 Available from: [in Polish]http://wwwold.pzh.gov.pl/oldpage/epimeld/index_p.html
62. Centers for Disease Control and Prevention (CDC): Early-season flu vaccination coverage - States November, 2018 Available from:https://www.cdc.gov/flu/fluvaxview/nifs-estimates-nov2018.htm
63. Kosaka M, Kotera Y, Tsuda K, Influenza vaccination uptake and attitudes among adult cancer patients in Japan: A web-based questionnaire survey before the 2020/2021 season: Hum Vaccin Immunother, 2021; 17(12); 5509-13
64. Chong Y, Matsumoto S, Kang D, Ikematsu H, Consecutive influenza surveillance of neuraminidase mutations and neuraminidase inhibitor resistance in Japan: Influenza Other Respir Viruses, 2019; 13; 115-22
65. Neuzil KM, Mellen BG, Wright PF, The effect of influenza on hospitalizations, outpatient visits, and courses of antibiotics in children: N Engl J Med, 2000; 342; 225-31
66. Buckley BS, Henschke N, Bergman H, Impact of vaccination on antibiotic usage: A systematic review and metaanalysis: Clin Microbiol Infect, 2019; 25; 1213-25
Figures
 Figure 1. Structure of neuraminidase A/Brisbane/02/2018 (A/H1N1/pdm09) with the R77G mutation detected in clinical sample number 145, as an example of a “viral oligomerization interface” mutation. The location of the mutation corresponds to position 77 of the B subunit chain (yellow backbone) and is 5 Ångström (A) from the A subunit oligomeric chain (blue backbone) in the A/H1N1/pdm09 neuraminidase gene (created using the FluSurver mutations app – https://gisaid.org/database-features/flusurver-mutations-app/).
Figure 1. Structure of neuraminidase A/Brisbane/02/2018 (A/H1N1/pdm09) with the R77G mutation detected in clinical sample number 145, as an example of a “viral oligomerization interface” mutation. The location of the mutation corresponds to position 77 of the B subunit chain (yellow backbone) and is 5 Ångström (A) from the A subunit oligomeric chain (blue backbone) in the A/H1N1/pdm09 neuraminidase gene (created using the FluSurver mutations app – https://gisaid.org/database-features/flusurver-mutations-app/). Figure 2. Structure of the neuraminidase of the virus B/Phuket/3073/2013 with the D356H mutation detected in clinical sample number 203, as an example of a mutation with the property of drug binding. The location of the mutation corresponds to position 224 of the A chain (yellow backbone) and is 5 Ångström (A) from the drug-binding site (created using the FluSurver mutations app – https://gisaid.org/database-features/flusurver-mutations-app/).
Figure 2. Structure of the neuraminidase of the virus B/Phuket/3073/2013 with the D356H mutation detected in clinical sample number 203, as an example of a mutation with the property of drug binding. The location of the mutation corresponds to position 224 of the A chain (yellow backbone) and is 5 Ångström (A) from the drug-binding site (created using the FluSurver mutations app – https://gisaid.org/database-features/flusurver-mutations-app/). Figure 3. Structure of the neuraminidase virus A/SingaporeINFIMH-16-0019/2016(A/H3N2/) with the D356H mutation detected in clinical sample number 25, as an example of a ligand-binding mutation. Mutation site 356 is shown in red on the A chain (yellow backbone), which is 5 Ångström (A) from the ligand-binding site (created using the FluSurver mutations app – https://gisaid.org/database-features/flusurver-mutations-app/).
Figure 3. Structure of the neuraminidase virus A/SingaporeINFIMH-16-0019/2016(A/H3N2/) with the D356H mutation detected in clinical sample number 25, as an example of a ligand-binding mutation. Mutation site 356 is shown in red on the A chain (yellow backbone), which is 5 Ångström (A) from the ligand-binding site (created using the FluSurver mutations app – https://gisaid.org/database-features/flusurver-mutations-app/). Figure 4. An example of a mutation site related to antibody-binding properties: the K369N mutation in the neuraminidase gene of virus A/Brisbane/02/2018 (A/H1N1/pdm09) in clinical sample number 770. The location of the mutation (red) corresponds to position 369 on the viral N chain (yellow backbone) and is 5 Ångström (A) from the heavy chain (H) antibodies (purple skeleton) (created using the FluSurver mutations app – https://gisaid.org/database-features/flusurver-mutations-app/).
Figure 4. An example of a mutation site related to antibody-binding properties: the K369N mutation in the neuraminidase gene of virus A/Brisbane/02/2018 (A/H1N1/pdm09) in clinical sample number 770. The location of the mutation (red) corresponds to position 369 on the viral N chain (yellow backbone) and is 5 Ångström (A) from the heavy chain (H) antibodies (purple skeleton) (created using the FluSurver mutations app – https://gisaid.org/database-features/flusurver-mutations-app/). Figure 5. Host protein-binding site exemplified by the G451R mutation in the neuraminidase gene of virus A/SingaporeINFIMH-16-0019/2016 (A/H3N2/) in clinical sample number 229. The location of the mutation corresponds to position 452 of the viral F chain (yellow backbone) of the neuraminidase gene and is 5 Ångström (A) from the D-chain of a non-viral protein (blue backbone), in this case the MHC class I antigen. The white backbone represents the other chains found in the structure (created using the FluSurver mutations app – https://gisaid.org/database-features/flusurver-mutations-app/).
Figure 5. Host protein-binding site exemplified by the G451R mutation in the neuraminidase gene of virus A/SingaporeINFIMH-16-0019/2016 (A/H3N2/) in clinical sample number 229. The location of the mutation corresponds to position 452 of the viral F chain (yellow backbone) of the neuraminidase gene and is 5 Ångström (A) from the D-chain of a non-viral protein (blue backbone), in this case the MHC class I antigen. The white backbone represents the other chains found in the structure (created using the FluSurver mutations app – https://gisaid.org/database-features/flusurver-mutations-app/). Figure 6. N-glycosylation on the example of the detected N329S mutation in the neuraminidase gene of virus A/SingaporeINFIMH-16-0019//2016 (A/H3N2/) in clinical sample number 30598. The mutation site corresponds to position 329 of the viral N chain (yellow backbone) and is located 5 Ångströms (A) from the ligand, the antibody heavy (H) chain (purple backbone), and the antibody light (L) chain (light purple backbone) (created using the FluSurver mutations app – https://gisaid.org/database-features/flusurver-mutations-app/).
Figure 6. N-glycosylation on the example of the detected N329S mutation in the neuraminidase gene of virus A/SingaporeINFIMH-16-0019//2016 (A/H3N2/) in clinical sample number 30598. The mutation site corresponds to position 329 of the viral N chain (yellow backbone) and is located 5 Ångströms (A) from the ligand, the antibody heavy (H) chain (purple backbone), and the antibody light (L) chain (light purple backbone) (created using the FluSurver mutations app – https://gisaid.org/database-features/flusurver-mutations-app/). Tables
 Table 1. Sequences of primers used in the RT-PCR for the detection of selected fragments of genes associated with resistance to anti-influenza drugs.
Table 1. Sequences of primers used in the RT-PCR for the detection of selected fragments of genes associated with resistance to anti-influenza drugs. Table 2. Summary of results related to the detected mutations in the influenza viruses tested.
Table 2. Summary of results related to the detected mutations in the influenza viruses tested. Table 3. Percentage distribution of other properties at the site of detected mutations in influenza viruses in Poland.
Table 3. Percentage distribution of other properties at the site of detected mutations in influenza viruses in Poland. Table 1. Sequences of primers used in the RT-PCR for the detection of selected fragments of genes associated with resistance to anti-influenza drugs.
Table 1. Sequences of primers used in the RT-PCR for the detection of selected fragments of genes associated with resistance to anti-influenza drugs. Table 2. Summary of results related to the detected mutations in the influenza viruses tested.
Table 2. Summary of results related to the detected mutations in the influenza viruses tested. Table 3. Percentage distribution of other properties at the site of detected mutations in influenza viruses in Poland.
Table 3. Percentage distribution of other properties at the site of detected mutations in influenza viruses in Poland. In Press
12 Mar 2024 : Clinical Research
Comparing Neuromuscular Blockade Measurement Between Upper Arm (TOF Cuff®) and Eyelid (TOF Scan®) Using Miv...Med Sci Monit In Press; DOI: 10.12659/MSM.943630
11 Mar 2024 : Clinical Research
Enhancement of Frozen-Thawed Human Sperm Quality with Zinc as a Cryoprotective AdditiveMed Sci Monit In Press; DOI: 10.12659/MSM.942946
12 Mar 2024 : Database Analysis
Risk Factors of Age-Related Macular Degeneration in a Population-Based Study: Results from SHIP-TREND-1 (St...Med Sci Monit In Press; DOI: 10.12659/MSM.943140
12 Mar 2024 : Clinical Research
Preoperative Blood Transfusion Requirements for Hemorrhoidal Severe Anemia: A Retrospective Study of 128 Pa...Med Sci Monit In Press; DOI: 10.12659/MSM.943126
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952








