18 December 2020: Database Analysis
Functional Analysis of Estrogen Receptor 1 in Diabetic Wound Healing: A Knockdown Cell-Based and Bioinformatic Study
Sha Qi1ABC, Qiong Han1BC, Danmou Xing1D, Long Qian1CE, Xiang Yu1E, Dong Ren1F, Huan Wang1E, Quan Chen1ABFG*DOI: 10.12659/MSM.928788
Med Sci Monit 2020; 26:e928788
Abstract
BACKGROUND: Diabetic wound (DW) treatment is a serious challenge for clinicians, and the underlying mechanisms of DWs remain elusive. We sought to identify the critical genes in the development of DWs and provide potential targets for DW therapies.
MATERIAL AND METHODS: Datasets of GSE38396 from the Gene Expression Omnibus (GEO) database were reviewed. Pathway analysis was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG), Gene Ontology term analyses were carried out, and Cytoscape software (Cytoscape 3.7.2) was used to construct the protein interaction network. Serum samples from patients with diabetes and control participants were collected, and the expression of estrogen receptor 1 (ESR1) was measured by quantitative reverse-transcription polymerase chain reaction. In addition, the function of ESR1 in human skin fibroblasts was investigated in vitro.
RESULTS: Eight samples were analyzed using the Morpheus online tool, which identified 637 upregulated and 448 downregulated differentially expressed genes. The top 5 KEGG pathways of upregulated differentially expressed genes were associated with sphingolipid metabolism, estrogen signaling, ECM-receptor interaction, MAPK signaling, and PI3K-Akt signaling. The hub genes for DWs were JUN, ESR1, CD44, SMARCA4, MMP2, BMP4, GSK3B, WDR5, PTK2, and PTGS2. JUN, MMP2, and ESR1 were the upregulated hub genes, and ESR1 was found to be consistently enriched in DW patients. Inhibition of ESR1 had a stimulative role in human skin fibroblasts.
CONCLUSIONS: ESR1 was identified as a crucial gene in the development of DWs, which suggests potential therapeutic targets for DW healing.
Keywords: Diabetic Angiopathies, Fibroblasts, Genes, vif, Databases, Genetic, Diabetes Complications, Estrogen Receptor alpha, Gene Expression Regulation, gene ontology, Skin Ulcer, Wound Healing
Background
It is widely accepted that a diagnosis of diabetes requires a substantial change in dietary habits and can produce overwhelming mental and social stress [1]. In addition, neurovascular lesions, secondary infection, hypoxia, and other harmful factors can cause diabetic wounds (DWs) that are challenging for clinicians to treat. Further, because these lesions are usually incurable, they cause economic and psychological burdens for patients [2]. A large number of studies have focused on the potential mechanisms underlying DW development [3]. For example, a recent study reported that circulating microRNA-20b-5p derived from the serum of patients with diabetes is associated with the deterioration of DWs, thereby providing a new therapeutic target for their treatment [4]. Similarly, a study regarding the therapeutic strategies for diabetic foot ulcers indicated that exosomal microRNA-15a-3p plays a crucial role in regulation of the healing progress through the NOX5/ROS signaling pathway [5]. Although many studies regarding DWs have been carried out, a clear mechanism for their development remains elusive.
Regulatory factors
A DNA microarray (also commonly known as DNA chip or biochip) is a collection of microscopic DNA spots attached to a solid surface. Such microarrays have been used to investigate the underlying pathogenic processes in a variety of diseases, and they have become crucial in functional genomic studies [8–10]. This technique has been widely applied to identify significant regulatory genes in diabetic diseases [11]. The differentially expressed genes (DEGs) are assumed to be related to changes in the levels of proteins that have significant functionality in this context and thus to have important regulatory effects in the progression of diabetes-related diseases. Therefore, we sought to identify critical DEGs in the development of DWs by using a comprehensive method based on bioinformatics.
Material and Methods
GENE CHIP DATA ACQUISITION:
The Gene Expression Omnibus (GEO;
IDENTIFICATION OF DEGS:
Morpheus (
ENRICHMENT ANALYSES OF GENE ONTOLOGY AND KYOTO ENCYCLOPEDIA OF GENES AND GENOMES PATHWAYS:
The online tool Database for Annotation, Visualization, and Integrated Discovery version (DAVID) Bioinformatics Resources 6.8 was used to the biological function of the genes [12,13]. Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed with DAVID.
PROTEIN–PROTEIN INTERACTION NETWORK CONSTRUCTION:
Upregulated and downregulated genes were imported into the software STRING (Search Tool for the Retrieval of Interacting Genes, https://string-db.org/), which identified the genes with a combined score of >0.5. (This score is based on the likelihood of interaction, and >0.5 is the default value.) Subsequently, the protein–protein interaction (PPI) network was constructed using Cytoscape software (version 3.7.2) [14]. Next, an analysis of the function enrichment was performed using DAVID, and the DEGs were ranked by degree centrality using the plugin software Centiscape 2.2 in Cytoscape. Degree represents the degree to which one node is associated with all other nodes in the network. Closeness represents how close a node is to other nodes in the network. Betweenness is the number of times that a node acts as the shortest bridge between 2 other nodes.
ETHICS APPROVAL:
The present study was approved by the Committees of Ethics in Wuhan Fourth Hospital (Wuhan, Hubei, China; 2019-021-17).
SAMPLE COLLECTION FROM PATIENTS AND HEALTHY CONTROLS:
From April 2019 to March 2020, peripheral blood samples from patients with DWs and healthy controls in our hospital (30 nondiabetic patients with foot trauma, and 30 diabetic patients with foot ulcers) were obtained for the subsequent validation of mRNA levels of the upregulated hub genes.
CELL CULTURE AND TRANSFECTION:
Human skin fibroblasts (HSFs; FuHeng Biology, Shanghai, China) were cultured in high-glucose Dulbecco’s modified Eagle’s medium (Gibco BRL, Grand Island, NY, USA) with 10% fetal bovine serum. For transfection, Lipofectamine 3000 (ThermoFisher Scientific) was used following the manufacturer’s instructions. Briefly, cells were cultured at 37°C with 5% CO2 and 95% humidity. For the transfection of mRNA and siRNA oligos steps, constructs from RIBOBIO (Guangzhou) were utilized, and cells were transfected with ESR1 siRNA (RIBOBIO, Guangzhou) at 50 nM.
QUANTITATIVE REVERSE-TRANSCRIPTION POLYMERASE CHAIN REACTION ANALYSES:
The total RNA was obtained from the serum samples. cDNA was then obtained through reverse and transcribe of the purified RNA. GAPDH served as an internal control. Relative miRNA expression levels were normalized to those of the internal control and were calculated according to the 2−ΔΔCq approach. The primer sequences were as follows:
CELL COUNTING KIT-8 MIGRATION ASSAY:
For the Cell Counting Kit-8 (CCK8) assay (Sigma, USA), HSFs (5×103 per well) were added to a 96-well plate, and cultured for 24, 48, or 72 h. Afterward, 20 μL of CCK-8 reagent was added into cells in serum-free medium for 2 h, followed by measurements of absorbance at 450 nm.
STATISTICAL ANALYSIS:
GraphPad Prism 8.0 was used to calculate the data, which are presented as mean±standard deviation. Paired data were compared using
Results
IDENTIFICATION OF DEGS:
The gene expression profile GSE38396 was obtained from the GEO database. There were 8 samples in this microarray data, which included 4 normal individuals and 4 patients with type 2 diabetes. The 8 samples were analyzed with the Morpheus online tool, which identified 637 upregulated and 448 downregulated DEGs. The top 30 upregulated genes and top 30 downregulated genes are shown in Figure 1.
GO AND KEGG ANALYSES:
GO analysis indicated that upregulated DEGs were significantly enriched in the following biological processes: positive regulation of GTPase activity, cardiovascular system development, circulatory system development, positive regulation of hydrolase activity, and heart development. The downregulated DEGs were enriched in cell proliferation, reproductive structure development, reproductive system development, negative regulation of cell proliferation, and negative regulation of macromolecule metabolic processes (Table 1). With regard to cellular components, upregulated DEGs were significantly enriched in endosome lumen, axoneme part, endoplasmic reticulum lumen, vacuolar lumen, and late endosome lumen, whereas the downregulated DEGs were enriched in nucleoplasm, nucleoplasm part, adherens junction, anchoring junction, and stress fiber (Table 2). Among molecular functions, upregulated DEGs were significantly enriched in guanyl-nucleotide exchange factor activity, molecular function regulator, Ras guanyl-nucleotide exchange factor activity, inositol phosphate phosphatase activity, and pyridoxal-phosphate binding, whereas the downregulated DEGs were enriched in poly(A) RNA binding, nucleotide binding, nucleoside phosphate binding, small molecule binding, and carbohydrate derivative binding (Table 3).
The top 5 KEGG pathways of upregulated DEGs were sphingolipid metabolism pathway, estrogen signaling pathway, ECM-receptor interaction pathway, MAPK signaling pathway, and PI3K-Akt signaling pathway. The top 5 KEGG pathways of downregulated DEGs were those for Hippo signaling, TGF-beta signaling, mRNA surveillance, AMPK signaling, and axon guidance (Table 4). The results of the KEGG enrichment analysis are shown in Figure 2.
PPI ANALYSIS:
The PPI network was obtained using STRING (https://string-db.org/cgi/input.pl) (Figure 3), and the top 10 hub genes with high degrees were then obtained using Cytoscape. The hub genes included JUN, ESR1, CD44, SMARCA4, MMP2, BMP4, GSK3B, WDR5, PTK2, and PTGS2 (Table 5). JUN had the highest degree, at 48. The expression boxplot of hub genes is shown in Figure 4. The degree, betweenness, and closeness of the hub genes are shown in Figure 5. The GO and KEGG pathway enrichment of the genes included in the PPI network showed that these genes were related to the biological processes of positive regulation of cell differentiation, cardiovascular system development, and circulatory system development; the molecular functions of molecular function regulator, transcription factor binding, and glycosaminoglycan binding; and the cellular components nucleoplasm, proteinaceous extracellular matrix, and contractile actin filament bundle. Most of the genes were enriched in the following signaling pathways: proteoglycans in the cancer pathway, pathways in cancer, and the Hippo signaling pathway (Table 6). The enrichment of the top 10 genes is shown in Figure 6. The expression of the top 100 genes from the PPI network and their chromosomal positions are shown in Figure 7.
:
The serum samples of the patients with DWs and the healthy controls were collected and quantitative real-time reverse-transcription polymerase chain reaction (qRT-PCR) analyses were performed to quantify ESR1 expression in these samples. As shown in Figure 8A, ESR1 was significantly upregulated in DW patients, which is consistent with the bioinformatic results. The effect of ESR1 on the proliferation of HSFs was then investigated in vitro. The CCK-8 assay showed that ESR1 inhibition promoted HSF proliferation (Figure 8B, 8C). Similarly, the qRT-PCR results indicated that the proliferation-related genes were increased and the apoptosis-related genes were decreased after ESR1 knockdown treatment (Figure 8D, 8E).
Discussion
Many studies have focused on the mechanisms underlying DWs, and the identification of the key genes involved in DW development is a crucial step in these investigations [15]. In the present study, we screened for key genes associated with DW development, using mRNA expression datasets from DW and control patients. This analysis led to the identification of
Matrix metalloprotein 2 (MMP2) has been reported to be involved in diverse functions, such as remodeling of the vasculature, angiogenesis, tissue repair, tumor invasion, inflammation, and atherosclerotic plaque rupture [20]. MMP2 was previously demonstrated to be involved in the regulation of wound healing, and the upregulation of MMP2 was reported to accelerate cutaneous wound healing via enhancing the functionality of keratinocytes [21]. Interestingly, MMP2 was also demonstrated to be associated with the development of diabetes [22]. In the present study, MMP2 was found to be upregulated in diabetes, which is consistent with a prior study [23]. Thus, we regard MMP2 as another potential key gene in DW development.
Estrogen receptor 1 (ESR1) is a type of nuclear hormone receptor. The steroid hormones and their receptors are involved in the regulation of eukaryotic gene expression and affect cellular proliferation and differentiation in target tissues. Ligand-dependent nuclear transactivation involves either direct homodimer binding to a palindromic estrogen response element sequence or association with other DNA-binding transcription factors, such as AP-1/c-Jun, c-Fos, ATF-2, Sp1, and Sp3, to mediate estrogen response element-independent signaling [24]. A prior study reported that ESR1 was related to the molecular mechanisms of astragalus membranaceus for treating type 2 diabetes mellitus [25]. Fibroblasts are the key cells in cutaneous wound repair, and their proper function is essential for wound contraction, collagen synthesis, and tissue remodeling. Therefore, we investigated how inhibition of ESR1 affects fibroblast function
Conclusions
In summary, the present study suggests that
Figures
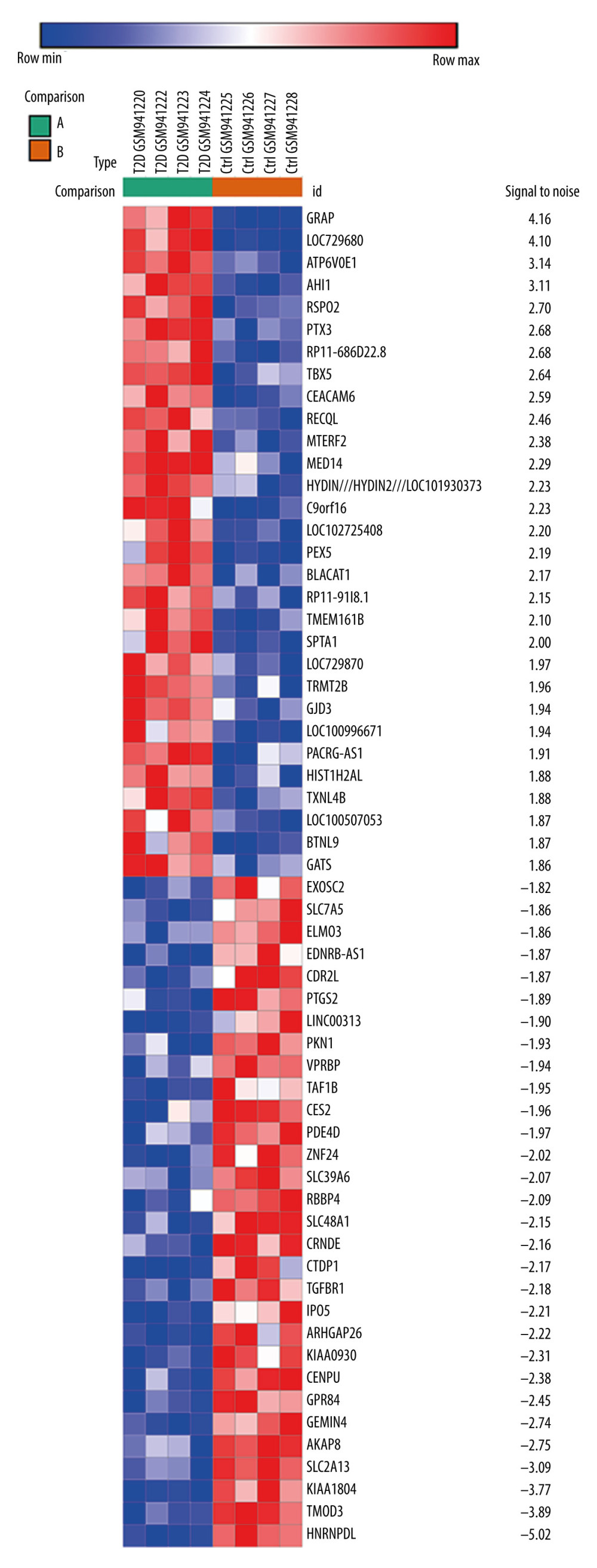 Figure 1. The top 60 differentially expressed genes (DEGs) of GSE38396 (30 upregulated and 30 downregulated) are presented in the heat map.
Figure 1. The top 60 differentially expressed genes (DEGs) of GSE38396 (30 upregulated and 30 downregulated) are presented in the heat map. 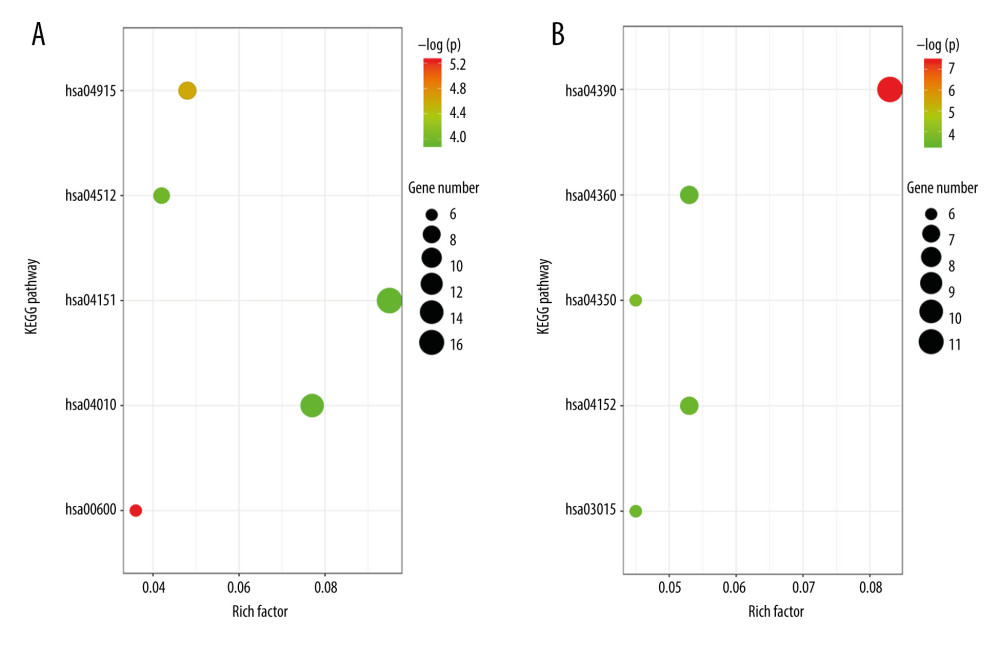 Figure 2. (A) Enrichment analysis results of upregulated genes: hsa00600, sphingolipid metabolism pathway; hsa04915, estrogen signaling pathway; hsa04512, ECM-receptor interaction pathway; hsa04010, MAPK signaling pathway; hsa04151, PI3K-Akt signaling pathway. (B) Enrichment analysis results of downregulated genes: hsa04390, Hippo signaling pathway; hsa04350, TGF-β signaling pathway; hsa03015, mRNA surveillance pathway; hsa04152, AMPK signaling pathway; hsa04360, Axon guidance pathway. ECM, extracellular matrix; MAPK, mitogen-activated protein kinase; PI3K-Akt, phosphatidylinositol-3-kinase and protein kinase B; TGF-β, transforming growth factor β.
Figure 2. (A) Enrichment analysis results of upregulated genes: hsa00600, sphingolipid metabolism pathway; hsa04915, estrogen signaling pathway; hsa04512, ECM-receptor interaction pathway; hsa04010, MAPK signaling pathway; hsa04151, PI3K-Akt signaling pathway. (B) Enrichment analysis results of downregulated genes: hsa04390, Hippo signaling pathway; hsa04350, TGF-β signaling pathway; hsa03015, mRNA surveillance pathway; hsa04152, AMPK signaling pathway; hsa04360, Axon guidance pathway. ECM, extracellular matrix; MAPK, mitogen-activated protein kinase; PI3K-Akt, phosphatidylinositol-3-kinase and protein kinase B; TGF-β, transforming growth factor β. 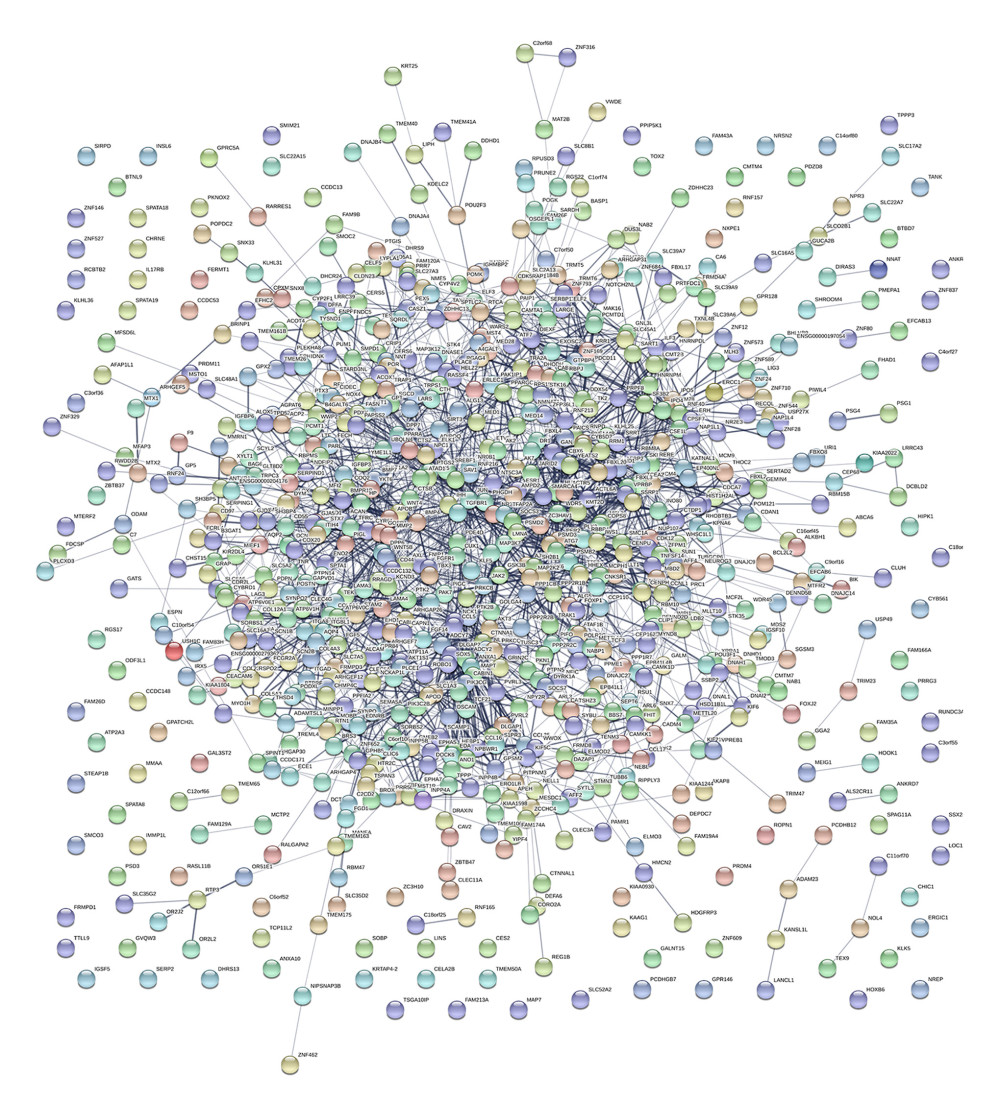 Figure 3. Interactions of the differentially expressed proteins. Protein–protein interaction (PPI) network constructed by STRING.
Figure 3. Interactions of the differentially expressed proteins. Protein–protein interaction (PPI) network constructed by STRING. 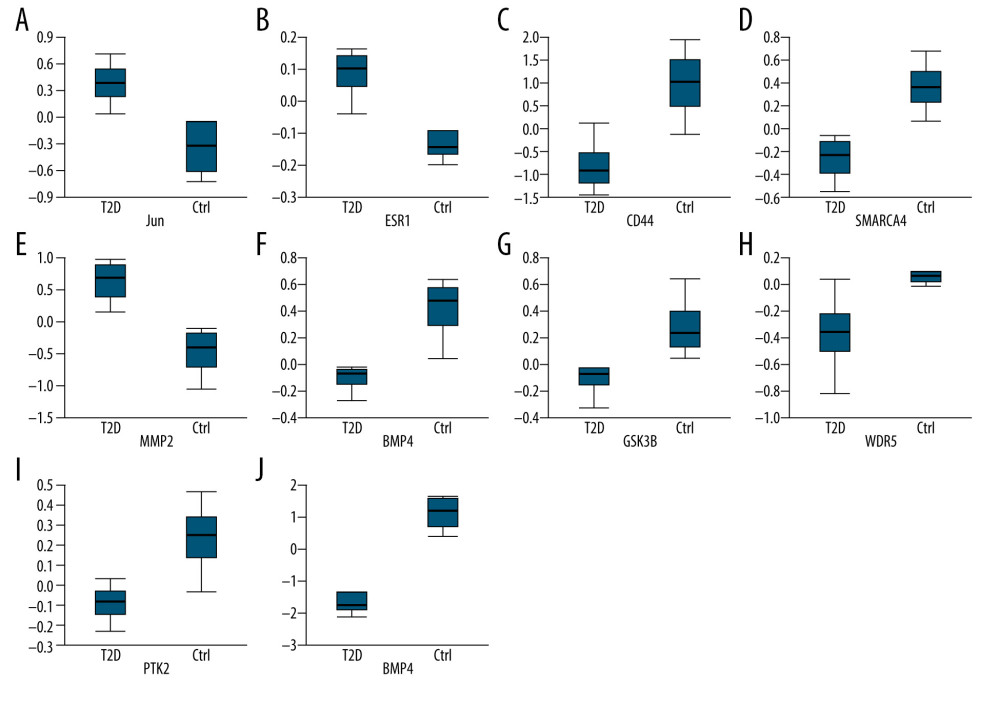 Figure 4. (A–J) Expression boxplot of JUN, ESR1, CD44, SMARCA4, MMP2, BMP4, GSK3B, WDR5, PTK2, and PTGS2 visualized by Morpheus.
Figure 4. (A–J) Expression boxplot of JUN, ESR1, CD44, SMARCA4, MMP2, BMP4, GSK3B, WDR5, PTK2, and PTGS2 visualized by Morpheus. 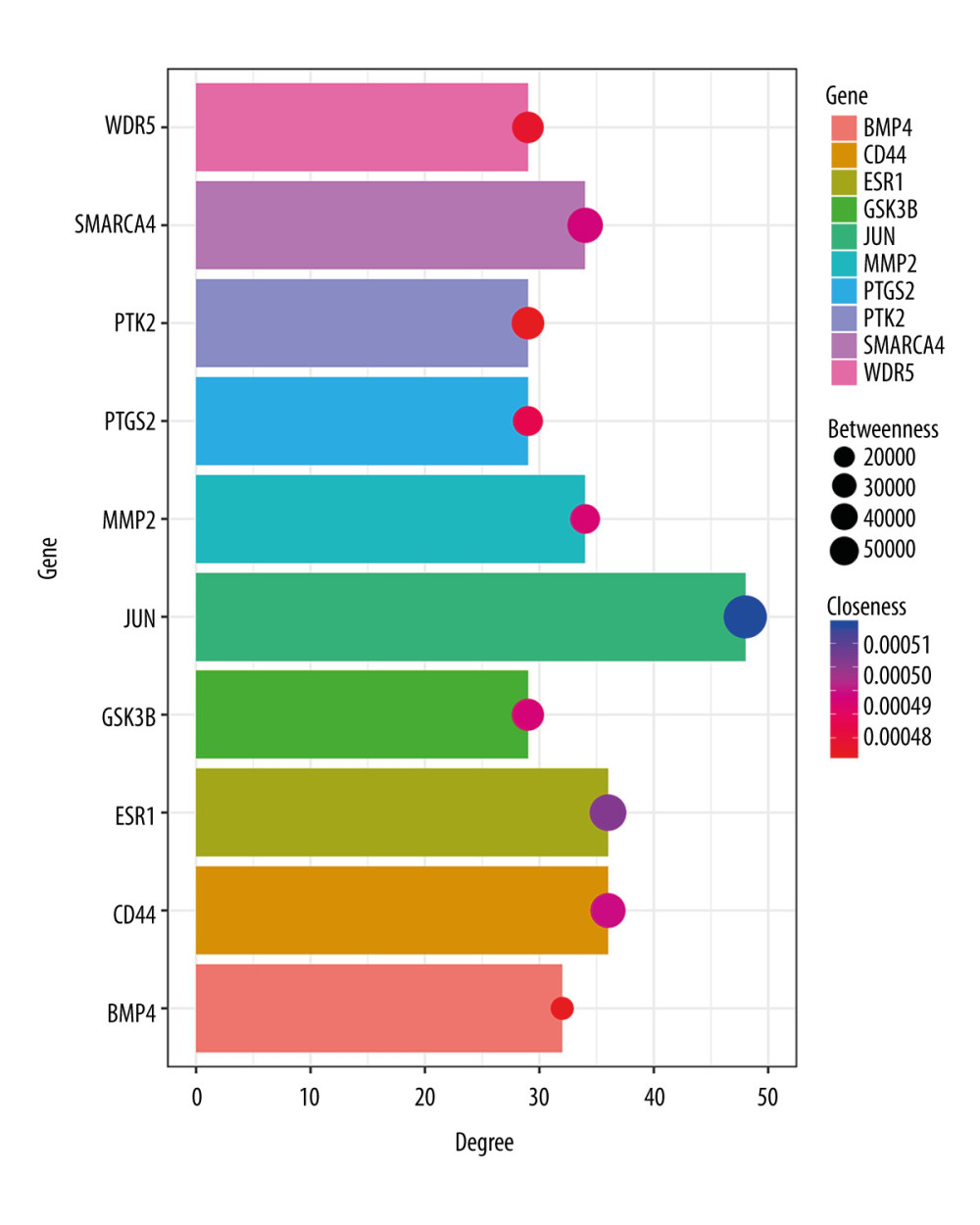 Figure 5. Degree, betweenness, and closeness of hub genes.
Figure 5. Degree, betweenness, and closeness of hub genes. 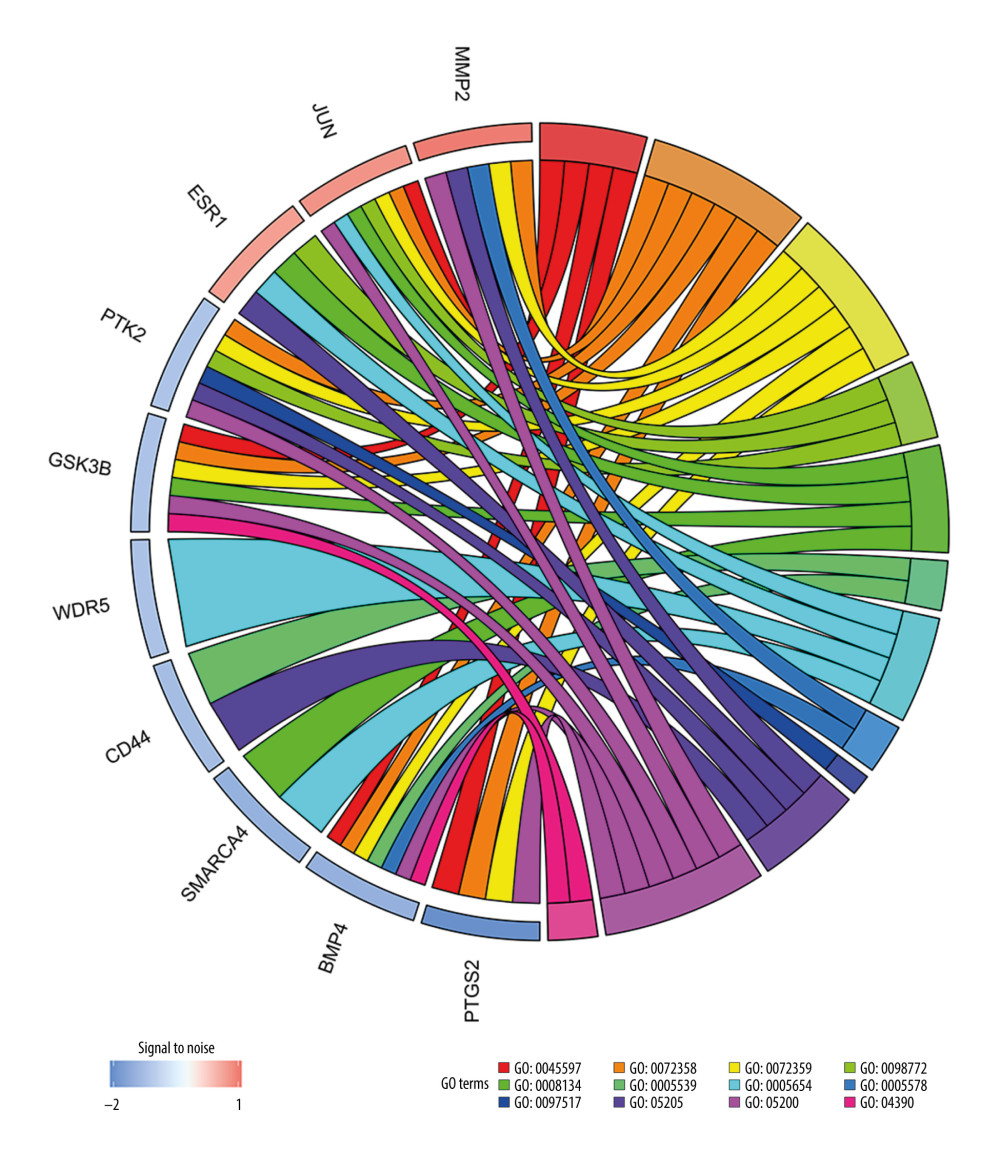 Figure 6. The enrichment of the top 10 genes in the protein–protein interaction (PPI) network. Biological processes: GO: 0045597, positive regulation of cell differentiation; GO: 0072358, cardiovascular system development; GO: 0072359, circulatory system development. Molecular functions: GO: 0098772, molecular function regulator; GO: 0008134, transcription factor binding; GO: 0005539, glycosaminoglycan binding. Cellular components: GO: 0005654, nucleoplasm; GO: 0005578, proteinaceous extracellular matrix; GO: 0097517, contractile actin filament bundle. KEGG pathway: hsa05205, proteoglycans in cancer pathway; hsa05200, pathways in cancer; hsa04390, Hippo signaling pathway.
Figure 6. The enrichment of the top 10 genes in the protein–protein interaction (PPI) network. Biological processes: GO: 0045597, positive regulation of cell differentiation; GO: 0072358, cardiovascular system development; GO: 0072359, circulatory system development. Molecular functions: GO: 0098772, molecular function regulator; GO: 0008134, transcription factor binding; GO: 0005539, glycosaminoglycan binding. Cellular components: GO: 0005654, nucleoplasm; GO: 0005578, proteinaceous extracellular matrix; GO: 0097517, contractile actin filament bundle. KEGG pathway: hsa05205, proteoglycans in cancer pathway; hsa05200, pathways in cancer; hsa04390, Hippo signaling pathway. 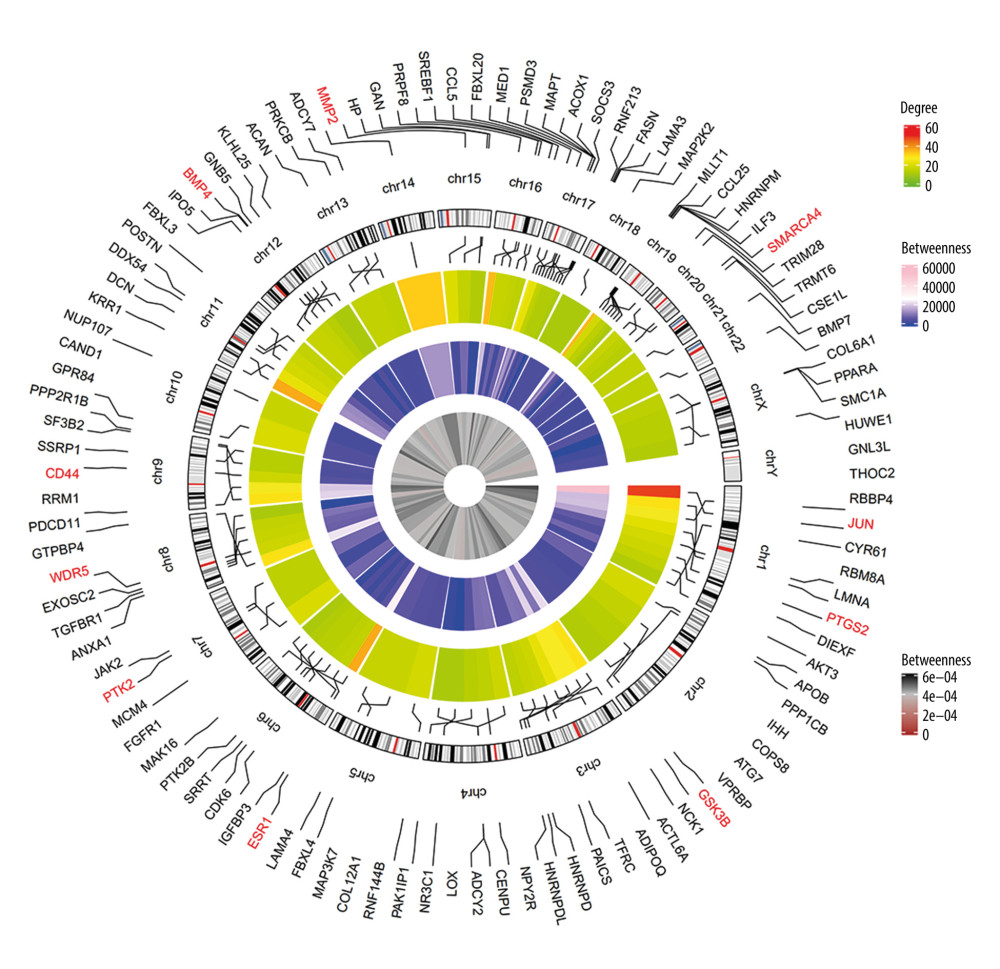 Figure 7. Circular visualization of chromosomal positions and connectivity of the top 100 genes in the protein–protein interaction (PPI) network. The names of the genes are shown in the outer circle. Different colors show different values of degree, betweenness, and closeness. The outer circle represents chromosomes; lines coming from each gene point to their specific chromosomal locations. The 10 hub genes are shown in red.
Figure 7. Circular visualization of chromosomal positions and connectivity of the top 100 genes in the protein–protein interaction (PPI) network. The names of the genes are shown in the outer circle. Different colors show different values of degree, betweenness, and closeness. The outer circle represents chromosomes; lines coming from each gene point to their specific chromosomal locations. The 10 hub genes are shown in red. 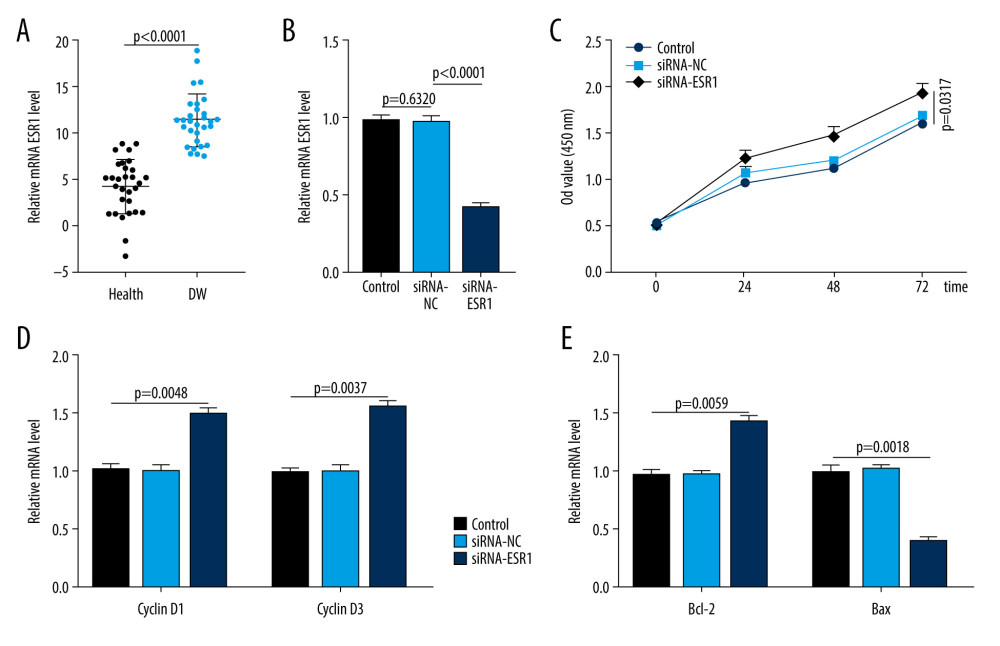 Figure 8. ESR1 is enriched in patients with diabetic wounds (DWs) and regulates human skin fibroblasts (HSFs). (A) The expression of ESR1 in the 2 groups was measured by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) analysis, n=30 per group. (B) The expression of ESR1 in cell groups with different treatments was measured by qRT-PCR analysis. (C) The CCK-8 results of HSFs in different groups. (D) The levels of cyclin D1 and cyclin D3 in different groups were measured by qRT-PCR analysis. (E) The levels of Bcl-2 and Bax were measured by qRT-PCR analysis.
Figure 8. ESR1 is enriched in patients with diabetic wounds (DWs) and regulates human skin fibroblasts (HSFs). (A) The expression of ESR1 in the 2 groups was measured by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) analysis, n=30 per group. (B) The expression of ESR1 in cell groups with different treatments was measured by qRT-PCR analysis. (C) The CCK-8 results of HSFs in different groups. (D) The levels of cyclin D1 and cyclin D3 in different groups were measured by qRT-PCR analysis. (E) The levels of Bcl-2 and Bax were measured by qRT-PCR analysis. Tables
Table 1. The upregulated and downregulated differentially expressed genes (DEGs) in biological process. Table 2. The upregulated and downregulated differentially expressed genes (DEGs) in cellular component.
Table 2. The upregulated and downregulated differentially expressed genes (DEGs) in cellular component.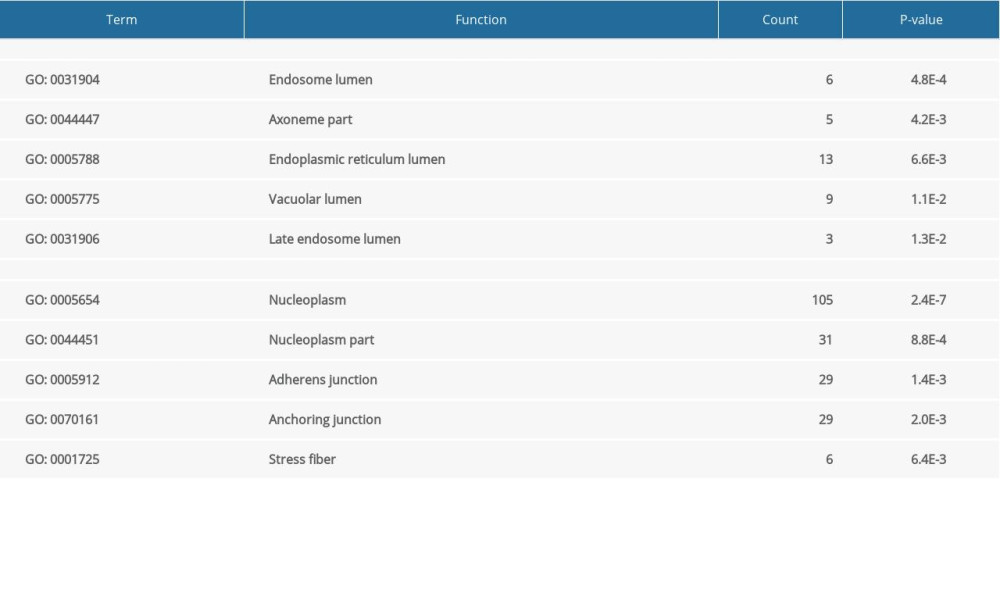 Table 3. The upregulated and downregulated differentially expressed genes (DEGs) in molecular function.
Table 3. The upregulated and downregulated differentially expressed genes (DEGs) in molecular function.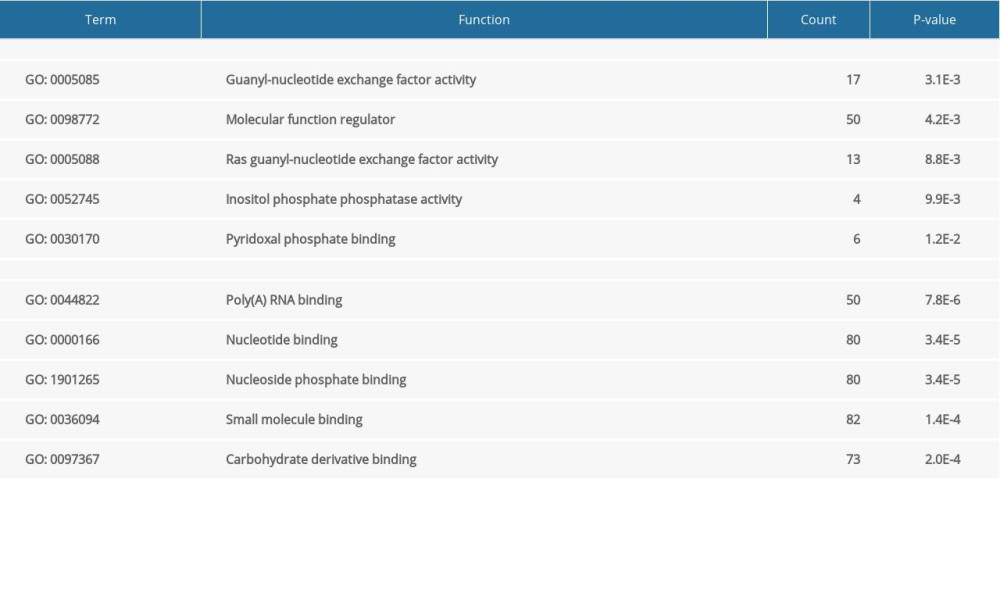 Table 4. The Kyoto Encyclopedia of Genes and Genomes analysis of upregulated and downregulated differentially expressed genes (DEGs).
Table 4. The Kyoto Encyclopedia of Genes and Genomes analysis of upregulated and downregulated differentially expressed genes (DEGs).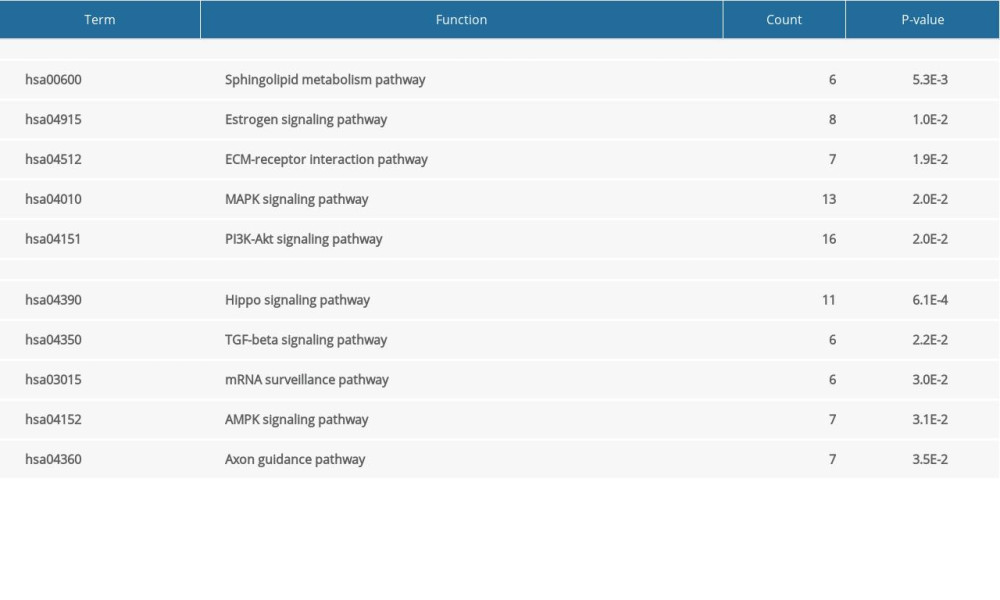 Table 5. The top 10 genes by degree of association between one node and other nodes.
Table 5. The top 10 genes by degree of association between one node and other nodes.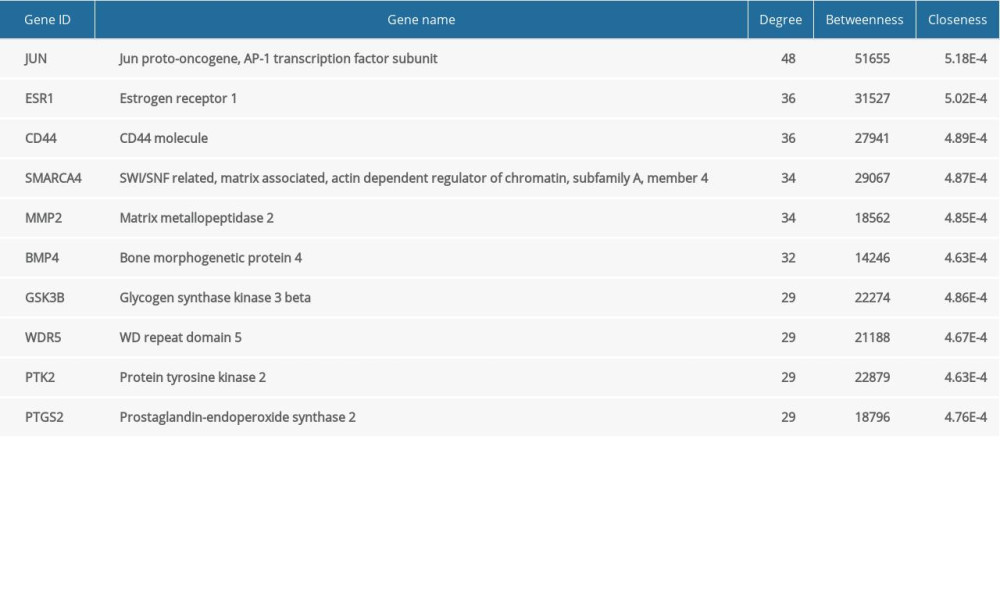 Table 6. Functional and pathway enrichment analyses of the genes in the protein–protein (PPI) network.
Table 6. Functional and pathway enrichment analyses of the genes in the protein–protein (PPI) network.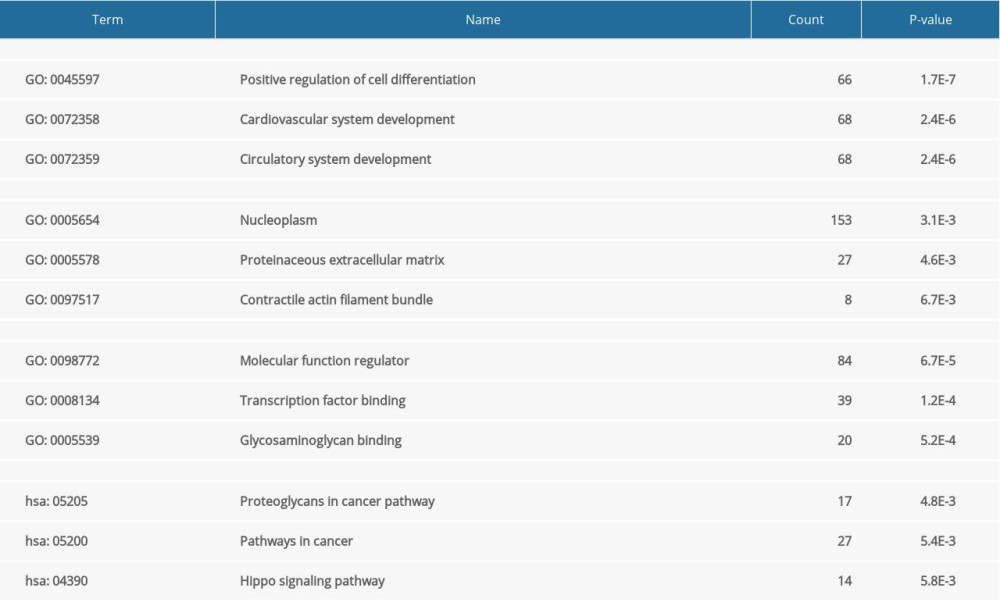
References
1. Argiana V, Kanellos PT, Eleftheriadou I, Low-glycemic-index/load desserts decrease glycemic and insulinemic response in patients with type 2 diabetes mellitus: Nutrients, 2020; 12(7); 2153
2. Xu Y, Cao K, Guo B, Lowered levels of nicotinic acetylcholine receptors and elevated apoptosis in the hippocampus of brains from patients with type 2 diabetes mellitus and db/db mice: Aging (Albany NY), 2020; 12(14); 14205-18
3. Pratley R, Dagogo-Jack S, Charbonnel B, Efficacy and safety of ertugliflozin in older patients with type 2 diabetes mellitus: A pooled analysis of phase III studies: Diabetes Obes Metab, 2020 [Online ahead of print]
4. Xiong Y, Chen L, Yan C, Circulating exosomal miR-20b-5p inhibition restores Wnt9b signaling and reverses diabetes-associated impaired wound healing: Small, 2020; 16(3); e1904044
5. Xiong Y, Chen L, Yu T, Inhibition of circulating exosomal microRNA-15a-3p accelerates diabetic wound repair: Aging (Albany NY), 2020; 12(10); 8968-86
6. Mi B, Chen L, Xiong Y, Saliva exosomes-derived UBE2O mRNA promotes angiogenesis in cutaneous wounds by targeting SMAD6: J Nanobiotechnology, 2020; 18(1); 68
7. Davis FM, denDekker A, Joshi AD, Palmitate-TLR4 signaling regulates the histone demethylase, JMJD3, in macrophages and impairs diabetic wound healing: Eur J Immunol, 2020 [Online ahead of print]
8. Xiong Y, Mi BB, Liu MF, Bioinformatics analysis and identification of genes and molecular pathways involved in synovial inflammation in rheumatoid arthritis: Med Sci Monit, 2019; 25; 2246-56
9. Yu T, You X, Zhou H, p53 plays a central role in the development of osteoporosis: Aging (Albany NY), 2020; 12(11); 10473-87
10. Yu T, Wang Z, You X, Resveratrol promotes osteogenesis and alleviates osteoporosis by inhibiting p53: Aging (Albany NY), 2020; 12(11); 10359-69
11. Xiong Y, Cao F, Chen L, Identification of key microRNAs and target genes for the diagnosis of bone nonunion: Mol Med Rep, 2020; 21(4); 1921-33
12. Huang DW, Sherman BT, Lempicki RA, Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists: Nucleic Acids Res, 2009; 37(1); 1-13
13. Huang DW, Sherman BT, Lempicki RA, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources: Nat Protoc, 2009; 4(1); 44-57
14. Shannon P, Markiel A, Ozier O, Cytoscape: A software environment for integrated models of biomolecular interaction networks: Genome Res, 2003; 13(11); 2498-504
15. Tian C, Ouyang X, Lv Q, Cross-talks between microRNAs and mRNAs in pancreatic tissues of streptozotocin-induced type 1 diabetic mice: Biomed Rep, 2015; 3(3); 333-42
16. Yun SJ, Jun KJ, Komori K, The regulation of TIM-3 transcription in T cells involves c-Jun binding but not CpG methylation at the TIM-3 promoter: Mol Immunol, 2016; 75; 60-68
17. Hu J, Li S, Sun X, Stk40 deletion elevates c-JUN protein level and impairs mesoderm differentiation: J Biol Chem, 2019; 294(25); 9959-72
18. Srivastava M, Saqib U, Banerjee S, Inhibition of the TIRAP-c-Jun interaction as a therapeutic strategy for AP1-mediated inflammatory responses: Int Immunopharmacol, 2019; 71; 188-97
19. Lu L, Ma J, Sun M, Melatonin ameliorates MI-induced cardiac remodeling and apoptosis through a JNK/p53-dependent mechanism in diabetes mellitus: Oxid Med Cell Longev, 2020; 2020 1535201
20. Diwanji N, Bergmann A, Basement membrane damage by ROS- and JNK-mediated Mmp2 activation drives macrophage recruitment to overgrown tissue: Nat Commun, 2020; 11(1); 3631
21. Zhang S, Zhu X, Li G, E2F1/SNHG7/miR-186-5p/MMP2 axis modulates the proliferation and migration of vascular endothelial cell in atherosclerosis: Life Sci, 2020; 257; 118013
22. Yang J, Zhao S, Tian F, SP1-mediated lncRNA PVT1 modulates the proliferation and apoptosis of lens epithelial cells in diabetic cataract via miR-214-3p/MMP2 axis: J Cell Mol Med, 2020; 24(1); 554-61
23. Nishihama K, Yasuma T, Yano Y, Anti-apoptotic activity of human matrix metalloproteinase-2 attenuates diabetes mellitus: Metabolism, 2018; 82; 88-99
24. Guclu-Geyik F, Coban N, Can G, Erginel-Unaltuna N, The rs2175898 polymorphism in the ESR1 gene has a significant sex-specific effect on obesity: Biochem Genet, 2020 [Online ahead of print]
25. Tan GC, Chu C, Lee YT, The influence of microsatellite polymorphisms in sex steroid receptor genes ESR1, ESR2 and AR on sex differences in brain structure: Neuroimage, 2020; 221; 117087
26. Moore K, Ghatnekar G, Gourdie RG, Potts JD, Impact of the controlled release of a connexin 43 peptide on corneal wound closure in an STZ model of type I diabetes: PLoS One, 2014; 9(1); e86570
Figures
 Figure 1. The top 60 differentially expressed genes (DEGs) of GSE38396 (30 upregulated and 30 downregulated) are presented in the heat map.
Figure 1. The top 60 differentially expressed genes (DEGs) of GSE38396 (30 upregulated and 30 downregulated) are presented in the heat map. Figure 2. (A) Enrichment analysis results of upregulated genes: hsa00600, sphingolipid metabolism pathway; hsa04915, estrogen signaling pathway; hsa04512, ECM-receptor interaction pathway; hsa04010, MAPK signaling pathway; hsa04151, PI3K-Akt signaling pathway. (B) Enrichment analysis results of downregulated genes: hsa04390, Hippo signaling pathway; hsa04350, TGF-β signaling pathway; hsa03015, mRNA surveillance pathway; hsa04152, AMPK signaling pathway; hsa04360, Axon guidance pathway. ECM, extracellular matrix; MAPK, mitogen-activated protein kinase; PI3K-Akt, phosphatidylinositol-3-kinase and protein kinase B; TGF-β, transforming growth factor β.
Figure 2. (A) Enrichment analysis results of upregulated genes: hsa00600, sphingolipid metabolism pathway; hsa04915, estrogen signaling pathway; hsa04512, ECM-receptor interaction pathway; hsa04010, MAPK signaling pathway; hsa04151, PI3K-Akt signaling pathway. (B) Enrichment analysis results of downregulated genes: hsa04390, Hippo signaling pathway; hsa04350, TGF-β signaling pathway; hsa03015, mRNA surveillance pathway; hsa04152, AMPK signaling pathway; hsa04360, Axon guidance pathway. ECM, extracellular matrix; MAPK, mitogen-activated protein kinase; PI3K-Akt, phosphatidylinositol-3-kinase and protein kinase B; TGF-β, transforming growth factor β. Figure 3. Interactions of the differentially expressed proteins. Protein–protein interaction (PPI) network constructed by STRING.
Figure 3. Interactions of the differentially expressed proteins. Protein–protein interaction (PPI) network constructed by STRING. Figure 4. (A–J) Expression boxplot of JUN, ESR1, CD44, SMARCA4, MMP2, BMP4, GSK3B, WDR5, PTK2, and PTGS2 visualized by Morpheus.
Figure 4. (A–J) Expression boxplot of JUN, ESR1, CD44, SMARCA4, MMP2, BMP4, GSK3B, WDR5, PTK2, and PTGS2 visualized by Morpheus. Figure 5. Degree, betweenness, and closeness of hub genes.
Figure 5. Degree, betweenness, and closeness of hub genes. Figure 6. The enrichment of the top 10 genes in the protein–protein interaction (PPI) network. Biological processes: GO: 0045597, positive regulation of cell differentiation; GO: 0072358, cardiovascular system development; GO: 0072359, circulatory system development. Molecular functions: GO: 0098772, molecular function regulator; GO: 0008134, transcription factor binding; GO: 0005539, glycosaminoglycan binding. Cellular components: GO: 0005654, nucleoplasm; GO: 0005578, proteinaceous extracellular matrix; GO: 0097517, contractile actin filament bundle. KEGG pathway: hsa05205, proteoglycans in cancer pathway; hsa05200, pathways in cancer; hsa04390, Hippo signaling pathway.
Figure 6. The enrichment of the top 10 genes in the protein–protein interaction (PPI) network. Biological processes: GO: 0045597, positive regulation of cell differentiation; GO: 0072358, cardiovascular system development; GO: 0072359, circulatory system development. Molecular functions: GO: 0098772, molecular function regulator; GO: 0008134, transcription factor binding; GO: 0005539, glycosaminoglycan binding. Cellular components: GO: 0005654, nucleoplasm; GO: 0005578, proteinaceous extracellular matrix; GO: 0097517, contractile actin filament bundle. KEGG pathway: hsa05205, proteoglycans in cancer pathway; hsa05200, pathways in cancer; hsa04390, Hippo signaling pathway. Figure 7. Circular visualization of chromosomal positions and connectivity of the top 100 genes in the protein–protein interaction (PPI) network. The names of the genes are shown in the outer circle. Different colors show different values of degree, betweenness, and closeness. The outer circle represents chromosomes; lines coming from each gene point to their specific chromosomal locations. The 10 hub genes are shown in red.
Figure 7. Circular visualization of chromosomal positions and connectivity of the top 100 genes in the protein–protein interaction (PPI) network. The names of the genes are shown in the outer circle. Different colors show different values of degree, betweenness, and closeness. The outer circle represents chromosomes; lines coming from each gene point to their specific chromosomal locations. The 10 hub genes are shown in red. Figure 8. ESR1 is enriched in patients with diabetic wounds (DWs) and regulates human skin fibroblasts (HSFs). (A) The expression of ESR1 in the 2 groups was measured by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) analysis, n=30 per group. (B) The expression of ESR1 in cell groups with different treatments was measured by qRT-PCR analysis. (C) The CCK-8 results of HSFs in different groups. (D) The levels of cyclin D1 and cyclin D3 in different groups were measured by qRT-PCR analysis. (E) The levels of Bcl-2 and Bax were measured by qRT-PCR analysis.
Figure 8. ESR1 is enriched in patients with diabetic wounds (DWs) and regulates human skin fibroblasts (HSFs). (A) The expression of ESR1 in the 2 groups was measured by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) analysis, n=30 per group. (B) The expression of ESR1 in cell groups with different treatments was measured by qRT-PCR analysis. (C) The CCK-8 results of HSFs in different groups. (D) The levels of cyclin D1 and cyclin D3 in different groups were measured by qRT-PCR analysis. (E) The levels of Bcl-2 and Bax were measured by qRT-PCR analysis. Tables
 Table 1. The upregulated and downregulated differentially expressed genes (DEGs) in biological process.
Table 1. The upregulated and downregulated differentially expressed genes (DEGs) in biological process. Table 2. The upregulated and downregulated differentially expressed genes (DEGs) in cellular component.
Table 2. The upregulated and downregulated differentially expressed genes (DEGs) in cellular component. Table 3. The upregulated and downregulated differentially expressed genes (DEGs) in molecular function.
Table 3. The upregulated and downregulated differentially expressed genes (DEGs) in molecular function. Table 4. The Kyoto Encyclopedia of Genes and Genomes analysis of upregulated and downregulated differentially expressed genes (DEGs).
Table 4. The Kyoto Encyclopedia of Genes and Genomes analysis of upregulated and downregulated differentially expressed genes (DEGs). Table 5. The top 10 genes by degree of association between one node and other nodes.
Table 5. The top 10 genes by degree of association between one node and other nodes. Table 6. Functional and pathway enrichment analyses of the genes in the protein–protein (PPI) network.
Table 6. Functional and pathway enrichment analyses of the genes in the protein–protein (PPI) network. Table 1. The upregulated and downregulated differentially expressed genes (DEGs) in biological process.
Table 1. The upregulated and downregulated differentially expressed genes (DEGs) in biological process. Table 2. The upregulated and downregulated differentially expressed genes (DEGs) in cellular component.
Table 2. The upregulated and downregulated differentially expressed genes (DEGs) in cellular component. Table 3. The upregulated and downregulated differentially expressed genes (DEGs) in molecular function.
Table 3. The upregulated and downregulated differentially expressed genes (DEGs) in molecular function. Table 4. The Kyoto Encyclopedia of Genes and Genomes analysis of upregulated and downregulated differentially expressed genes (DEGs).
Table 4. The Kyoto Encyclopedia of Genes and Genomes analysis of upregulated and downregulated differentially expressed genes (DEGs). Table 5. The top 10 genes by degree of association between one node and other nodes.
Table 5. The top 10 genes by degree of association between one node and other nodes. Table 6. Functional and pathway enrichment analyses of the genes in the protein–protein (PPI) network.
Table 6. Functional and pathway enrichment analyses of the genes in the protein–protein (PPI) network. In Press
08 Mar 2024 : Clinical Research
Evaluation of Foot Structure in Preschool Children Based on Body MassMed Sci Monit In Press; DOI: 10.12659/MSM.943765
15 Apr 2024 : Laboratory Research
The Role of Copper-Induced M2 Macrophage Polarization in Protecting Cartilage Matrix in OsteoarthritisMed Sci Monit In Press; DOI: 10.12659/MSM.943738
07 Mar 2024 : Clinical Research
Knowledge of and Attitudes Toward Clinical Trials: A Questionnaire-Based Study of 179 Male Third- and Fourt...Med Sci Monit In Press; DOI: 10.12659/MSM.943468
08 Mar 2024 : Animal Research
Modification of Experimental Model of Necrotizing Enterocolitis (NEC) in Rat Pups by Single Exposure to Hyp...Med Sci Monit In Press; DOI: 10.12659/MSM.943443
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952








